Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity - PubMed (original) (raw)
Review
Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity
Emmanuelle Charpentier et al. FEMS Microbiol Rev. 2015 May.
Abstract
CRISPR-Cas is an RNA-mediated adaptive immune system that defends bacteria and archaea against mobile genetic elements. Short mature CRISPR RNAs (crRNAs) are key elements in the interference step of the immune pathway. A CRISPR array composed of a series of repeats interspaced by spacer sequences acquired from invading mobile genomes is transcribed as a precursor crRNA (pre-crRNA) molecule. This pre-crRNA undergoes one or two maturation steps to generate the mature crRNAs that guide CRISPR-associated (Cas) protein(s) to cognate invading genomes for their destruction. Different types of CRISPR-Cas systems have evolved distinct crRNA biogenesis pathways that implicate highly sophisticated processing mechanisms. In Types I and III CRISPR-Cas systems, a specific endoribonuclease of the Cas6 family, either standalone or in a complex with other Cas proteins, cleaves the pre-crRNA within the repeat regions. In Type II systems, the trans-acting small RNA (tracrRNA) base pairs with each repeat of the pre-crRNA to form a dual-RNA that is cleaved by the housekeeping RNase III in the presence of the protein Cas9. In this review, we present a detailed comparative analysis of pre-crRNA recognition and cleavage mechanisms involved in the biogenesis of guide crRNAs in the three CRISPR-Cas types.
Keywords: Cas5d; Cas6; Cas9; RNase III; crRNA biogenesis; tracrRNA.
© FEMS 2015.
Figures
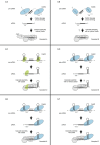
Figure 1.
cas gene composition of the CRISPR-Cas systems. Loci from Types I-A to I-F, Types II-A to II-C and Types III-A and II-B CRISPR-Cas systems are represented. The CRISPR arrays are composed of a series of repeats (black diamonds) interspaced by invading genome-targeting spacers (colored diamonds). An operon of cas genes is located in the close vicinity of the CRISPR array. The Cas proteins involved in the crRNA biogenesis in Types I-A, I-B, I-D, I-E and I-F and Types III-A and III-B belong to the Cas6 family. An exception is the gene product Cas5d responsible for the processing of pre-crRNA in Type I-C. In Type II systems, tracrRNA, and the proteins Cas9 and RNase III are the three components responsible for pre-crRNA maturation.
Figure 2.
crRNA processing pathways in Type I CRISPR-Cas systems. In Type I systems, the palindromic repeats in the pre-crRNA are either unstructured (Cascade/I-A, Cascade/I-B) or form hairpin structures (Cascade/I-C, Cascade/I-D, Cascade/I-E, Cascade/I-F) that are recognized by the nuclease Cas6 (Cas6a, Cascade/I-A; Cas6b, Cascade/I-B; Cas6d, Cascade/I-D; Cas6e, Cascade/I-E; Cas6f, Cascade/I-F) or Cas5 (Cas5d, Cascade/I-C). After cleavage, the crRNA hairpin remains associated with Cas6 or Cas5 whilst other subunits bind the 5′ handle and spacer, which is used for the recognition of cognate genetic element sequences by the respective Cascade complexes.
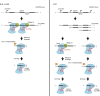
Figure 3.
crRNA processing pathways in Type II CRISPR-Cas systems. In Type II systems, the precursor transcript of the CRISPR repeat-spacer array forms duplexes with the _trans_-activating tracrRNA through pre-crRNA repeat:tracrRNA anti-repeat interactions. The duplex RNAs stabilized by the protein Cas9 are recognized and cleaved by the bacterial endoribonuclease III (RNase III). A second processing by unknown nucleases (trimming by an exonuclease and/or cleavage by an endoribonuclease) generates the mature crRNAs. An alternative pathway for the production of mature crRNAs was described in a Type II-C of N. meningitidis. Here, the transcription of short crRNAs occurs directly from promoters contained within the repeats of the array, and thus independently of cleavage by RNase III. The mature dual tracrRNA:crRNAs complexed with the protein Cas9 form the interference complex that target and cleave site specifically double-stranded DNA.
Figure 4.
crRNA processing pathways in Type III CRISPR-Cas systems. In Type III-A and III-B systems, the standalone Cas6 endonuclease binds unstructured pre-crRNA and cleaves within each repeat to generate intermediate crRNAs with 5′ and 3′ repeat-derived termini. The crRNAs are loaded into the Csm (Type III-A) or Cmr (Type III-B) complex and undergo further maturation through trimming of the 3′ repeat-derived sequence by nucleases that are yet to be identified.
Similar articles
- The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA.
Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. Fonfara I, et al. Nature. 2016 Apr 28;532(7600):517-21. doi: 10.1038/nature17945. Epub 2016 Apr 20. Nature. 2016. PMID: 27096362 - Approaches to study CRISPR RNA biogenesis and the key players involved.
Behler J, Hess WR. Behler J, et al. Methods. 2020 Feb 1;172:12-26. doi: 10.1016/j.ymeth.2019.07.015. Epub 2019 Jul 17. Methods. 2020. PMID: 31325492 Review. - The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems.
Chylinski K, Le Rhun A, Charpentier E. Chylinski K, et al. RNA Biol. 2013 May;10(5):726-37. doi: 10.4161/rna.24321. Epub 2013 Apr 5. RNA Biol. 2013. PMID: 23563642 Free PMC article. - A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii.
Brendel J, Stoll B, Lange SJ, Sharma K, Lenz C, Stachler AE, Maier LK, Richter H, Nickel L, Schmitz RA, Randau L, Allers T, Urlaub H, Backofen R, Marchfelder A. Brendel J, et al. J Biol Chem. 2014 Mar 7;289(10):7164-7177. doi: 10.1074/jbc.M113.508184. Epub 2014 Jan 23. J Biol Chem. 2014. PMID: 24459147 Free PMC article. - CRISPR-Cas systems and RNA-guided interference.
Barrangou R. Barrangou R. Wiley Interdiscip Rev RNA. 2013 May-Jun;4(3):267-78. doi: 10.1002/wrna.1159. Epub 2013 Mar 20. Wiley Interdiscip Rev RNA. 2013. PMID: 23520078 Review.
Cited by
- Targeting ABCB1-mediated tumor multidrug resistance by CRISPR/Cas9-based genome editing.
Yang Y, Qiu JG, Li Y, Di JM, Zhang WJ, Jiang QW, Zheng DW, Chen Y, Wei MN, Huang JR, Wang K, Shi Z. Yang Y, et al. Am J Transl Res. 2016 Sep 15;8(9):3986-3994. eCollection 2016. Am J Transl Res. 2016. PMID: 27725879 Free PMC article. - DNA Targeting by a Minimal CRISPR RNA-Guided Cascade.
Hochstrasser ML, Taylor DW, Kornfeld JE, Nogales E, Doudna JA. Hochstrasser ML, et al. Mol Cell. 2016 Sep 1;63(5):840-51. doi: 10.1016/j.molcel.2016.07.027. Mol Cell. 2016. PMID: 27588603 Free PMC article. - Genetic advancements in obesity management and CRISPR-Cas9-based gene editing system.
Jayachandran M, Fei Z, Qu S. Jayachandran M, et al. Mol Cell Biochem. 2023 Mar;478(3):491-501. doi: 10.1007/s11010-022-04518-w. Epub 2022 Jul 31. Mol Cell Biochem. 2023. PMID: 35909208 Review. - Classification of CRISPR/Cas system and its application in tomato breeding.
Chaudhuri A, Halder K, Datta A. Chaudhuri A, et al. Theor Appl Genet. 2022 Feb;135(2):367-387. doi: 10.1007/s00122-021-03984-y. Epub 2022 Jan 1. Theor Appl Genet. 2022. PMID: 34973111 Free PMC article. Review. - Global View of Candidate Therapeutic Target Genes in Hormone-Responsive Breast Cancer.
Salvati A, Gigantino V, Nassa G, Mirici Cappa V, Ventola GM, Cracas DGC, Mastrocinque R, Rizzo F, Tarallo R, Weisz A, Giurato G. Salvati A, et al. Int J Mol Sci. 2020 Jun 6;21(11):4068. doi: 10.3390/ijms21114068. Int J Mol Sci. 2020. PMID: 32517194 Free PMC article. Review.
References
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. - PubMed
- Briner AE, Donohoue PD, Gomaa AA, et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources