ClinGen--the Clinical Genome Resource - PubMed (original) (raw)
. 2015 Jun 4;372(23):2235-42.
doi: 10.1056/NEJMsr1406261. Epub 2015 May 27.
Jonathan S Berg, Lisa D Brooks, Carlos D Bustamante, James P Evans, Melissa J Landrum, David H Ledbetter, Donna R Maglott, Christa Lese Martin, Robert L Nussbaum, Sharon E Plon, Erin M Ramos, Stephen T Sherry, Michael S Watson; ClinGen
Collaborators, Affiliations
- PMID: 26014595
- PMCID: PMC4474187
- DOI: 10.1056/NEJMsr1406261
ClinGen--the Clinical Genome Resource
Heidi L Rehm et al. N Engl J Med. 2015.
Abstract
On autopsy, a patient is found to have hypertrophic cardiomyopathy. The patient’s family pursues genetic testing that shows a “likely pathogenic” variant for the condition on the basis of a study in an original research publication. Given the dominant inheritance of the condition and the risk of sudden cardiac death, other family members are tested for the genetic variant to determine their risk. Several family members test negative and are told that they are not at risk for hypertrophic cardiomyopathy and sudden cardiac death, and those who test positive are told that they need to be regularly monitored for cardiomyopathy on echocardiography. Five years later, during a routine clinic visit of one of the genotype-positive family members, the cardiologist queries a database for current knowledge on the genetic variant and discovers that the variant is now interpreted as “likely benign” by another laboratory that uses more recently derived population-frequency data. A newly available testing panel for additional genes that are implicated in hypertrophic cardiomyopathy is initiated on an affected family member, and a different variant is found that is determined to be pathogenic. Family members are retested, and one member who previously tested negative is now found to be positive for this new variant. An immediate clinical workup detects evidence of cardiomyopathy, and an intracardiac defibrillator is implanted to reduce the risk of sudden cardiac death.
Figures
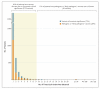
Figure 1. Variant Histogram from Mendelian Disease Testing of 15,000 Probands
Shown are data for 5839 variants that have been found in patients with cardiomyopathy, hearing loss, RASopathies (i.e., developmental syndromes caused by germline mutations that alter the Ras subfamily), aortopathies, hereditary cancers, pulmonary disorders, skin disorders, and other genetic syndromes, as tested by the Laboratory for Molecular Medicine at Partners HealthCare. Shown at the top of the chart are the percentages of patients who carry frequently observed pathogenic variants and patients who have variants that are rare or of uncertain significance.
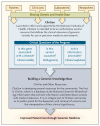
Figure 2. Clinical Genome Resource (ClinGen)
More information on ClinGen is available at
, and more information on ClinVar is available at
.
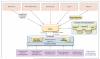
Figure 3. Flow of Data through ClinGen
Shown is the typical flow of information into ClinVar and ClinGenKB, a new database that is designed to allow for a flexible working environment for curation. Most variants are submitted by external sources and databases directly into ClinVar for immediate access by the community. Variants then flow into ClinGenKB to enable the resolution of differences in interpretation, as well as expert review of variants by the clinical-domain working groups that are shown. Additional sources of data and machine-learning algorithms may be brought into ClinGenKB to aid in the interpretive process. BIC denotes Breast Cancer Information Core, CFTR2 Clinical and Functional Translation of CFTR, InSiGHT the variant database for the International Society for Gastrointestinal Hereditary Tumours, OMIM Online Mendelian Inheritance in Man, and PharmGKB the Pharmacogenomics Knowledge Base.
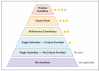
Figure 4. Review Levels Annotated in ClinVar
Variants with assertions are rated according to the source and level of review for each submitted variant assertion. Submitters must comply with requirements (
www.ncbi.nlm.nih.gov/clinvar/docs/assertion\_criteria
) for a submission to be assigned one, three, or four stars. Two stars are automatically assigned when multiple one-star submitted assertions are consistent. The distinction between submitters that have provided criteria and those that have not will begin in June 2015.
Comment in
- ClinGen and Genetic Testing.
Karam R, Pesaran T, Chao E. Karam R, et al. N Engl J Med. 2015 Oct;373(14):1376-7. doi: 10.1056/NEJMc1508700. N Engl J Med. 2015. PMID: 26422737 No abstract available. - ClinGen and Genetic Testing.
Nussbaum RL, Rehm HL; ClinGen. Nussbaum RL, et al. N Engl J Med. 2015 Oct;373(14):1379. doi: 10.1056/NEJMc1508700. N Engl J Med. 2015. PMID: 26430707 No abstract available.
Similar articles
- ClinGen and Genetic Testing.
Nussbaum RL, Rehm HL; ClinGen. Nussbaum RL, et al. N Engl J Med. 2015 Oct;373(14):1379. doi: 10.1056/NEJMc1508700. N Engl J Med. 2015. PMID: 26430707 No abstract available. - ClinGen and Genetic Testing.
Karam R, Pesaran T, Chao E. Karam R, et al. N Engl J Med. 2015 Oct;373(14):1376-7. doi: 10.1056/NEJMc1508700. N Engl J Med. 2015. PMID: 26422737 No abstract available. - ClinGen and Genetic Testing.
Easton DF, Pharoah PD, Foulkes WD. Easton DF, et al. N Engl J Med. 2015 Oct;373(14):1378. doi: 10.1056/NEJMc1508700. N Engl J Med. 2015. PMID: 26422736 No abstract available. - Detecting Causal Variants in Mendelian Disorders Using Whole-Genome Sequencing.
Hamzeh AR, Andrews TD, Field MA. Hamzeh AR, et al. Methods Mol Biol. 2021;2243:1-25. doi: 10.1007/978-1-0716-1103-6_1. Methods Mol Biol. 2021. PMID: 33606250 Review. - Genetic Testing in Inherited Heart Diseases.
Ingles J, Macciocca I, Morales A, Thomson K. Ingles J, et al. Heart Lung Circ. 2020 Apr;29(4):505-511. doi: 10.1016/j.hlc.2019.10.014. Epub 2019 Nov 29. Heart Lung Circ. 2020. PMID: 31813745 Review.
Cited by
- Elevated plasma miR-210 expression is associated with atypical genitalia in patients with 46,XY differences in sex development.
Elias FM, Nishi MY, Sircili MHP, Bastista RL, Gomes NL, Ferrari MTM, Costa EMF, Denes FT, Mendonca BB, Domenice S. Elias FM, et al. Mol Genet Genomic Med. 2022 Dec;10(12):e2084. doi: 10.1002/mgg3.2084. Epub 2022 Nov 11. Mol Genet Genomic Med. 2022. PMID: 36369742 Free PMC article. - Towards precision medicine.
Ashley EA. Ashley EA. Nat Rev Genet. 2016 Aug 16;17(9):507-22. doi: 10.1038/nrg.2016.86. Nat Rev Genet. 2016. PMID: 27528417 Review. - Deep generative modeling of the human proteome reveals over a hundred novel genes involved in rare genetic disorders.
Orenbuch R, Kollasch AW, Spinner HD, Shearer CA, Hopf TA, Franceschi D, Dias M, Frazer J, Marks DS. Orenbuch R, et al. Res Sq [Preprint]. 2024 Jan 4:rs.3.rs-3740259. doi: 10.21203/rs.3.rs-3740259/v1. Res Sq. 2024. PMID: 38260496 Free PMC article. Preprint. - ATAV: a comprehensive platform for population-scale genomic analyses.
Ren Z, Povysil G, Hostyk JA, Cui H, Bhardwaj N, Goldstein DB. Ren Z, et al. BMC Bioinformatics. 2021 Mar 23;22(1):149. doi: 10.1186/s12859-021-04071-1. BMC Bioinformatics. 2021. PMID: 33757430 Free PMC article. - All for one and one for all: heterogeneity of genetic etiologies in neurodevelopmental psychiatric disorders.
Moreno-De-Luca D, Martin CL. Moreno-De-Luca D, et al. Curr Opin Genet Dev. 2021 Jun;68:71-78. doi: 10.1016/j.gde.2021.02.015. Epub 2021 Mar 25. Curr Opin Genet Dev. 2021. PMID: 33773394 Free PMC article. Review.
References
- McKusick-Nathans Institute of Genetic Medicine. Johns Hopkins Medicine Online Mendelian Inheritance in Man (OMIM) http://omim.org.
- 1000 Genomes Project 1000 Genomes: a deep catalog of human genetic variation. http://www.1000genomes.org.
- National Research Council . Sharing publication-related data and materials: responsibilities of authorship in the life sciences. National Academies Press; Washington, DC: 2003. - PubMed
- Chanock SJ, Manolio T, Boehnke M, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–60. - PubMed
Publication types
MeSH terms
Grants and funding
- U41 HG006834/HG/NHGRI NIH HHS/United States
- T15 LM007124/LM/NLM NIH HHS/United States
- P30 ES013508/ES/NIEHS NIH HHS/United States
- U01 HG007436/HG/NHGRI NIH HHS/United States
- U41 HG009650/HG/NHGRI NIH HHS/United States
- U01 HG007437/HG/NHGRI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- Z99 LM999999/ImNIH/Intramural NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases