Complete Bypass of Restriction Systems for Major Staphylococcus aureus Lineages - PubMed (original) (raw)
Complete Bypass of Restriction Systems for Major Staphylococcus aureus Lineages
Ian R Monk et al. mBio. 2015.
Erratum in
- Erratum for Monk et al., Complete Bypass of Restriction Systems for Major Staphylococcus aureus Lineages.
Monk IR, Tree JJ, Howden BP, Stinear TP, Foster TJ. Monk IR, et al. mBio. 2016 Mar 1;7(2):e00171. doi: 10.1128/mBio.00171-16. mBio. 2016. PMID: 26933053 Free PMC article. No abstract available.
Abstract
Staphylococcus aureus is a prominent global nosocomial and community-acquired bacterial pathogen. A strong restriction barrier presents a major hurdle for the introduction of recombinant DNA into clinical isolates of S. aureus. Here, we describe the construction and characterization of the IMXXB series of Escherichia coli strains that mimic the type I adenine methylation profiles of S. aureus clonal complexes 1, 8, 30, and ST93. The IMXXB strains enable direct, high-efficiency transformation and streamlined genetic manipulation of major S. aureus lineages.
Importance: The genetic manipulation of clinical S. aureus isolates has been hampered due to the presence of restriction modification barriers that detect and subsequently degrade inappropriately methylated DNA. Current methods allow the introduction of plasmid DNA into a limited subset of S. aureus strains at high efficiency after passage of plasmid DNA through the restriction-negative, modification-proficient strain RN4220. Here, we have constructed and validated a suite of E. coli strains that mimic the adenine methylation profiles of different clonal complexes and show high-efficiency plasmid DNA transfer. The ability to bypass RN4220 will reduce the cost and time involved for plasmid transfer into S. aureus. The IMXXB series of E. coli strains should expedite the process of mutant construction in diverse genetic backgrounds and allow the application of new techniques to the genetic manipulation of S. aureus.
Copyright © 2015 Monk et al.
Figures
FIG 1
SMRT sequencing for the identification of adenine methylation in S. aureus and engineered E. coli. (A) The target recognition motifs (TRMs) for the type I HsdMS systems encoded by each S. aureus strain were identified, and the positions of methylated adenine residues on the chromosome plotted with Circos (34). Each adenine methylation on the chromosome is represented by a line whose length corresponds with the interpulse duration of the read. (B and C) PacBio reads were analyzed by the SMRT suite pipeline version 2.2.0/motif finder 1.3.1 to identify conserved adenine-methylated residues (in boldface) and the TRMs for the HsdS alleles of each S. aureus (B) and IMXXB E. coli (C) strain.1, Mean modification QV is defined as the quality value of the base calls within the motif.2, Mean motif coverage is defined as the average depth of read coverage within a motif.3, GATC methylation encoded by dam in the E. coli strain is not present in S. aureus.
FIG 2
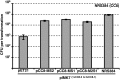
Transformation of NRS384 with HsdMS-methylated plasmid. Three different CC8 HsdMS combinations were expressed from an IPTG-inducible plasmid (pET21) in E. coli BL21 (including the hybrid _hsdM2_-hsdS1). The coextracted shuttle vector pIMK7 (5 µg total plasmid DNA) was transformed into CA-MRSA strain NRS384. pIMK7 isolated from NRS384 was included as the maximal transformation of fully modified plasmid. The number of TRMs on pIMK7 for CC8 strains are indicated next to the plasmid name. The transformation efficiencies are expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows data representative of the data from three independent experiments.
FIG 3
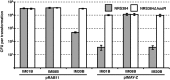
Construction and characterization of IMXXB E. coli strains. (A) Schematic of the construction of an E. coli strain expressing CC-specific hsdMS alleles from strong promoters at neutral locations in the DC10B (DH10B Δ_dcm_) chromosome. (B to D) Transformation profiles of S. aureus strains (grey bars) and their isogenic hsdR mutants (defective in type I restriction) (white bars) with plasmid pRAB11 (5 µg) or pIMAY-Z (2.5 µg) isolated from DC10B or the respective IMXXB strain of E. coli. The number of TRMs on either plasmid for the CC of the strain is denoted next to the plasmid name. NT, no transformants were detected. The transformation efficiencies are expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows data representative of the data from three independent experiments.
FIG 4
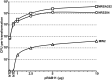
Effect of plasmid concentration for S. aureus electroporation. Different concentrations of plasmid DNA (pRAB11 at 0.1, 0.5, 1, 2.5, 5, and 10 µg) were isolated from E. coli strains IM01B, IM08B, and IM30B for transformation into S. aureus MW2, NRS384, and MRSA252, respectively. A dose-dependent response was observed for up to 5 µg of plasmid with pRAB11. The transformation efficiencies are expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows representative data from one experiment.
FIG 5
Determination of the TRM for each TRD. Clustal Omega alignment of HsdS proteins from CC1, CC5, CC8, CC30, and ST93. The protein sequences of the HsdS variants were aligned with Clustal Omega, and TRDs that match exactly are color coded in the same color.
FIG 6
Transformation of JKD6159 and type I restriction mutants with pRAB11. Plasmid pRAB11 was isolated from E. coli strains DC10B and IM93B and S. aureus strain JKD6159 for transformation into JKD6159 and the respective type I-deficient mutants. pRAB11 contains 4 TRM sites in ST93-1, 2 in ST93-2, and 2 in ST93-3. The transformation efficiencies were expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows representative data from one experiment. NT, no transformants were isolated.
FIG 7
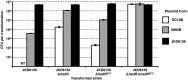
Transformation of S. aureus NRS384 and NRS384 ΔhsdR with plasmid isolated from IMXXB E. coli. Plasmid pRAB11 or pIMAY-Z was isolated from E. coli strains IM01B, IM08B, and IM30B for transformation into a CC8 host (NRS384) and the respective hsdR mutant. The transformation efficiencies are expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows representative data from one experiment.
FIG 8
Transformation of additional representative isolates from CC1, CC8/ST239, and CC30 with plasmid isolated from IMXXB strains. Plasmid pRAB11 (5 µg) was isolated from either E. coli strain DC10B or the compatible IMXXB strain for transformation into additional representative S. aureus isolates of CC1, CC8/ST239, and CC30. The transformation efficiencies are expressed as the mean numbers of all transformants obtained in each experiment ± standard deviations (error bars) from three replicates. The graph shows representative data from one experiment.
Similar articles
- Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis.
Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. Monk IR, et al. mBio. 2012 Mar 20;3(2):e00277-11. doi: 10.1128/mBio.00277-11. Print 2012. mBio. 2012. PMID: 22434850 Free PMC article. - Inactivation of the SauI type I restriction-modification system is not sufficient to generate Staphylococcus aureus strains capable of efficiently accepting foreign DNA.
Veiga H, Pinho MG. Veiga H, et al. Appl Environ Microbiol. 2009 May;75(10):3034-8. doi: 10.1128/AEM.01862-08. Epub 2009 Mar 20. Appl Environ Microbiol. 2009. PMID: 19304835 Free PMC article. - Genetic manipulation of Staphylococci-breaking through the barrier.
Monk IR, Foster TJ. Monk IR, et al. Front Cell Infect Microbiol. 2012 Apr 12;2:49. doi: 10.3389/fcimb.2012.00049. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919640 Free PMC article. Review. - Efficient Genome Editing in Most Staphylococcus aureus by Using the Restriction-Modification System Silent CRISPR-Cas9 Toolkit.
Yang P, Yang J, Lin T, Liu Q, Yin Y, Chen D, Yang S. Yang P, et al. ACS Synth Biol. 2023 Nov 17;12(11):3340-3351. doi: 10.1021/acssynbio.3c00339. Epub 2023 Oct 13. ACS Synth Biol. 2023. PMID: 37830328 - Genomic variation and evolution of Staphylococcus aureus.
Lindsay JA. Lindsay JA. Int J Med Microbiol. 2010 Feb;300(2-3):98-103. doi: 10.1016/j.ijmm.2009.08.013. Epub 2009 Oct 7. Int J Med Microbiol. 2010. PMID: 19811948 Review.
Cited by
- Molecular Characterization of Clinical Rel Mutations and Consequences for Resistance Expression and Fitness in Staphylococcus aureus.
Deventer AT, Bryson D, Shortill M, Boraston AB, Hobbs JK. Deventer AT, et al. Antimicrob Agents Chemother. 2022 Dec 20;66(12):e0093822. doi: 10.1128/aac.00938-22. Epub 2022 Nov 8. Antimicrob Agents Chemother. 2022. PMID: 36346240 Free PMC article. - Genetic variation of staphylococcal LukAB toxin determines receptor tropism.
Perelman SS, James DBA, Boguslawski KM, Nelson CW, Ilmain JK, Zwack EE, Prescott RA, Mohamed A, Tam K, Chan R, Narechania A, Pawline MB, Vozhilla N, Moustafa AM, Kim SY, Dittmann M, Ekiert DC, Bhabha G, Shopsin B, Planet PJ, Koralov SB, Torres VJ. Perelman SS, et al. Nat Microbiol. 2021 Jun;6(6):731-745. doi: 10.1038/s41564-021-00890-3. Epub 2021 Apr 19. Nat Microbiol. 2021. PMID: 33875847 Free PMC article. - Development of an inducer-free, virulence gene promoter-controlled, and fluorescent reporter-labeled CRISPR interference system in Staphylococcus aureus.
Miah R, Johannessen M, Kjos M, Lentz CS. Miah R, et al. Microbiol Spectr. 2024 Oct 3;12(10):e0060224. doi: 10.1128/spectrum.00602-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162514 Free PMC article. - Clinical Mutations That Partially Activate the Stringent Response Confer Multidrug Tolerance in Staphylococcus aureus.
Bryson D, Hettle AG, Boraston AB, Hobbs JK. Bryson D, et al. Antimicrob Agents Chemother. 2020 Feb 21;64(3):e02103-19. doi: 10.1128/AAC.02103-19. Print 2020 Feb 21. Antimicrob Agents Chemother. 2020. PMID: 31871080 Free PMC article. - Unstable chromosome rearrangements in Staphylococcus aureus cause phenotype switching associated with persistent infections.
Guérillot R, Kostoulias X, Donovan L, Li L, Carter GP, Hachani A, Vandelannoote K, Giulieri S, Monk IR, Kunimoto M, Starrs L, Burgio G, Seemann T, Peleg AY, Stinear TP, Howden BP. Guérillot R, et al. Proc Natl Acad Sci U S A. 2019 Oct 1;116(40):20135-20140. doi: 10.1073/pnas.1904861116. Epub 2019 Sep 16. Proc Natl Acad Sci U S A. 2019. PMID: 31527262 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials