A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage - PubMed (original) (raw)
A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage
Marjolein van Sluis et al. Genes Dev. 2015.
Abstract
DNA double-strand breaks (DSBs) are repaired by two main pathways: nonhomologous end-joining and homologous recombination (HR). Repair pathway choice is thought to be determined by cell cycle timing and chromatin context. Nucleoli, prominent nuclear subdomains and sites of ribosome biogenesis, form around nucleolar organizer regions (NORs) that contain rDNA arrays located on human acrocentric chromosome p-arms. Actively transcribed rDNA repeats are positioned within the interior of the nucleolus, whereas sequences proximal and distal to NORs are packaged as heterochromatin located at the nucleolar periphery. NORs provide an opportunity to investigate the DSB response at highly transcribed, repetitive, and essential loci. Targeted introduction of DSBs into rDNA, but not abutting sequences, results in ATM-dependent inhibition of their transcription by RNA polymerase I. This is coupled with movement of rDNA from the nucleolar interior to anchoring points at the periphery. Reorganization renders rDNA accessible to repair factors normally excluded from nucleoli. Importantly, DSBs within rDNA recruit the HR machinery throughout the cell cycle. Additionally, unscheduled DNA synthesis, consistent with HR at damaged NORs, can be observed in G1 cells. These results suggest that HR can be templated in cis and suggest a role for chromosomal context in the maintenance of NOR genomic stability.
Keywords: DNA double-strand breaks (DSBs); ataxia telangiectasia-mutated (ATM); homologous recombination (HR); nucleolar organizer region (NOR); nucleolus; ribosomal genes (rDNA).
© 2015 van Sluis and McStay; Published by Cold Spring Harbor Laboratory Press.
Figures
Figure 1.
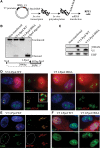
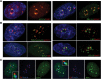
Introduction of DSBs into rDNA with I-PpoI induces nucleolar reorganization and inhibition of transcription. (A) Strategy for expressing I-PpoI. A transcript encoding V5 epitope-tagged I-PpoI preceded by an IRES element was produced in vitro using T7 RNA polymerase. Following polyadenylation, transcripts were transfected into RPE1 cells. (B) Southern blotting reveals that wild-type (WT) I-PpoI introduces DSBs within the 28S rRNA-coding sequence in vivo. Approximately 20% of rDNA repeats contain a DSB. Genomic DNA digested to completion with I-PpoI in vitro is included for comparison. Note the absence of cleavage observed using catalytically dead I-PpoI (H98A). The probing scheme is shown below. (C) Western blotting with V5 antibodies reveals that wild-type and mutant I-PpoI are produced at comparable levels and that only wild-type I-PpoI induces a γH2AX response. (D) Staining of cells 6 h after transfection reveals that wild-type but not mutant V5-tagged I-PpoI induces formation of UBF-containing nucleolar caps that are associated with prominent γH2AX signals. The inset shows an enlargement of a single nucleolus. (E) BrUTP incorporation assays indicate that transcription of rDNA by Pol I is inhibited in wild-type I-PpoI transfected cells. The nucleus of the transfected cell shown is indicated by a white dotted line. (F) EU incorporation assays confirm that wild-type but not mutant V5-tagged I-PpoI induces inhibition of nucleolar (Pol I) transcription and further show that nucleoplasmic (Pol II and III) transcription is unaffected.
Figure 2.
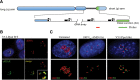
I-PpoI-induced nucleolar caps contain rDNA and form adjacent to DJ sequences. (A) A cartoon representation of a human acrocentric chromosome with enlargement of the rDNA array and surrounding sequences. Sequences immediately distal (DJ) to the rDNA are colored green. The position of probes used in FISH to detect DJs (green) and rDNA (red) are indicated below. (B) 3D immunoFISH performed on I-PpoI transfected cells shows colocalization of rDNA and γH2AX in nucleolar caps. The inset shows an enlarged individual nucleolus. (C) 3D immunoFISH performed on AMD-treated and I-PpoI transfected cells shows that, in both cases, nucleolar caps form immediately adjacent to DJ sequences embedded in perinucleolar heterochromatin.
Figure 3.
Introduction of DSBs into rDNA and adjacent sequences using CRISPR/Cas9 reveals a regional specificity of response. (A) Cas9 complexed with gRNAs targeted to rDNA induces a γH2AX response, nucleolar segregation, and inhibition of nucleolar transcription. The locations of gRNA targets within the rDNA repeat are illustrated. In all cases, transfected cells are identified with a Cas9 antibody. (Top three panels) Nucleolar segregation was assayed by UBF staining. Ongoing transcription was assessed by EU incorporation. The middle three panels represent the majority of cells in which nucleolar transcription is absent. The bottom three panels represent the minority of transfected cells in which partial inhibition was observed. (B) Cas9 complexed with DJ targeted gRNAs induces a nucleolar γH2AX response but does not result in nucleolar segregation or inhibition of transcription. The positions of DJ gRNAs are illustrated. Nucleolar segregation and ongoing transcription are shown in the top and bottom panels respectively.
Figure 4.
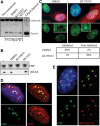
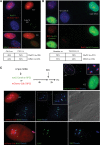
Transcriptional inhibition is ATM-dependent. (A) Southern blotting reveals that ATM and DNA-PK inhibitors (10 μM KU55933 and 10 μM NU7441, respectively) have little effect on the cleavage efficiency of I-PpoI in vivo. (B) Western blotting indicates that ATM inhibitor (KU55933) selectively inhibits the γH2AX response induced by I-PpoI transfection. (C) Nucleolar transcription continues in V5 I-PpoI transfected cells in the presence of ATM inhibitor (KU55933). Nucleolar transcription is inhibited in control (DMSO-treated) cells. Quantification of results from analyzing >100 transfected cells for each treatment are shown below. The insets show enlarged single nucleoli. (D) Staining of I-PpoI transfected cells with an ATM antibody shows enrichment of ATM at nucleolar caps. (E) Staining of I-PpoI transfected cells with antibodies that detect ATM-pS1981 show that activated ATM is restricted to nucleolar caps.
Figure 5.
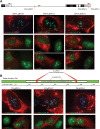
I-PpoI-induced DSBs selectively recruit HR repair factors to nucleolar caps. (A) BRCA1 and 53BP1 were individually observed at γH2AX-positive nucleolar caps in I-PpoI transfected cells. (B) Antibody staining of I-PpoI transfected cells shows that HR factors RPA2 (total and phospho-S4/S8), Rad51, and Rad52 are highly enriched at nucleolar caps. (C) 3D immunoFISH was performed on I-PpoI transfected RPE1 cells using a DJ probe, Treacle, and RPA2 antibodies. The insets show enlarged individual caps containing a single NOR.
Figure 6.
HR factors are recruited, and damage-induced DNA synthesis can be observed at I-PpoI-induced nucleolar caps in G1 cells. (A) I-PpoI transfection of RPE1 cells stably expressing FUCCI component mCherry-Cdt1 reveals that HR factors Rad51 and RPA2 are recruited comparably to nucleolar caps in G1 (red) and late S/G2 (nonred) cells. Quantification is shown below. (B) I-PpoI transfection of RPE1 cells stably expressing FUCCI component mAG-Geminin reveals that HR factors Rad51 and RPA2 are recruited comparably to nucleolar caps in G1 (nongreen) and late S/G2 (green) cells. Quantification is shown below. (C) RPE1 cells stably expressing FUCCI component mAG-Geminin or mCherry-Cdt1 were transfected with I-PpoI mRNA; 4 h after transfection, fresh medium containing EdU was added to cells. Following a 2-h pulse of EdU labeling, cells were fixed. Treacle antibodies detected damage-induced nucleolar caps. Damage-induced DNA synthesis was visualized using biotin-azide and fluorophore-conjugated streptavidin (see the Materials and Methods for details). In the top right panel, EdU incorporation can be observed at damage-induced nucleolar caps in G1 (nongreen) mAG-Geminin cells. In the middle panels, EdU incorporation is observed in nucleolar caps in both G1 (red) and G2 (nonred) mCherry-Cdt1 cells. A DIC image is shown at the right. In the bottom panels, a single enlarged G1 cell from the same experiment is shown.
Figure 7.
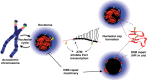
A model for chromosomal context influencing genomic stability of rDNA arrays. rDNA arrays on acrocentric short arms are surrounded by sequences that form perinucleolar heterochromatin during interphase. DJ sequences (green) are localized within this heterochromatin, providing an anchor for the linked rDNA array (red). Introduction of DSBs (yellow) into the rDNA induce ATM-dependent inhibition of rDNA transcription by Pol I followed by subsequent nucleolar reorganization. Positioning of rDNA within caps on the nucleolar surface is a consequence of anchoring by DJ sequences. We hypothesize that this nucleolar reorganization renders damaged rDNA accessible to DSB repair factors normally excluded from nucleoli. Concentration of damaged and intact rDNA repeats within nucleolar caps promotes repair by the HR pathway. As this occurs in G1 cells, HR repair may be templated by repeats in cis within the same NOR. See the Discussion for further details.
Similar articles
- NORs on human acrocentric chromosome p-arms are active by default and can associate with nucleoli independently of rDNA.
van Sluis M, van Vuuren C, Mangan H, McStay B. van Sluis M, et al. Proc Natl Acad Sci U S A. 2020 May 12;117(19):10368-10377. doi: 10.1073/pnas.2001812117. Epub 2020 Apr 24. Proc Natl Acad Sci U S A. 2020. PMID: 32332163 Free PMC article. - Nucleolar reorganization in response to rDNA damage.
van Sluis M, McStay B. van Sluis M, et al. Curr Opin Cell Biol. 2017 Jun;46:81-86. doi: 10.1016/j.ceb.2017.03.004. Epub 2017 Apr 18. Curr Opin Cell Biol. 2017. PMID: 28431265 Review. - Nucleolar DNA Double-Strand Break Responses Underpinning rDNA Genomic Stability.
van Sluis M, McStay B. van Sluis M, et al. Trends Genet. 2019 Oct;35(10):743-753. doi: 10.1016/j.tig.2019.07.001. Epub 2019 Jul 25. Trends Genet. 2019. PMID: 31353047 Review. - Nucleolar organizer regions: genomic 'dark matter' requiring illumination.
McStay B. McStay B. Genes Dev. 2016 Jul 15;30(14):1598-610. doi: 10.1101/gad.283838.116. Genes Dev. 2016. PMID: 27474438 Free PMC article. Review. - Double-strand breaks in ribosomal RNA genes activate a distinct signaling and chromatin response to facilitate nucleolar restructuring and repair.
Korsholm LM, Gál Z, Lin L, Quevedo O, Ahmad DA, Dulina E, Luo Y, Bartek J, Larsen DH. Korsholm LM, et al. Nucleic Acids Res. 2019 Sep 5;47(15):8019-8035. doi: 10.1093/nar/gkz518. Nucleic Acids Res. 2019. PMID: 31184714 Free PMC article.
Cited by
- DNA repair: location, location, location.
van Sluis M, McStay B. van Sluis M, et al. Oncotarget. 2015 Jul 10;6(19):16828-9. doi: 10.18632/oncotarget.4840. Oncotarget. 2015. PMID: 26219556 Free PMC article. No abstract available. - The Nucleolus and PARP1 in Cancer Biology.
Engbrecht M, Mangerich A. Engbrecht M, et al. Cancers (Basel). 2020 Jul 6;12(7):1813. doi: 10.3390/cancers12071813. Cancers (Basel). 2020. PMID: 32640701 Free PMC article. Review. - Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly.
Li X, Tyler JK. Li X, et al. Elife. 2016 Jun 8;5:e15129. doi: 10.7554/eLife.15129. Elife. 2016. PMID: 27269284 Free PMC article. - Treacle is Upregulated in Cancer and Correlates With Poor Prognosis.
Oxe KC, Larsen DH. Oxe KC, et al. Front Cell Dev Biol. 2022 Jun 20;10:918544. doi: 10.3389/fcell.2022.918544. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35794866 Free PMC article. - NORs on human acrocentric chromosome p-arms are active by default and can associate with nucleoli independently of rDNA.
van Sluis M, van Vuuren C, Mangan H, McStay B. van Sluis M, et al. Proc Natl Acad Sci U S A. 2020 May 12;117(19):10368-10377. doi: 10.1073/pnas.2001812117. Epub 2020 Apr 24. Proc Natl Acad Sci U S A. 2020. PMID: 32332163 Free PMC article.
References
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. 2002. Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11. - PubMed
- Berkovich E, Monnat RJ Jr, Kastan MB. 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9: 683–690. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous