Ordered Regions of Channel Nucleoporins Nup62, Nup54, and Nup58 Form Dynamic Complexes in Solution - PubMed (original) (raw)
Ordered Regions of Channel Nucleoporins Nup62, Nup54, and Nup58 Form Dynamic Complexes in Solution
Alok Sharma et al. J Biol Chem. 2015.
Abstract
Three out of ∼30 nucleoporins, Nup62, Nup54, and Nup58, line the nuclear pore channel. These "channel" nucleoporins each contain an ordered region of ∼150-200 residues, which is predicted to be segmented into 3-4 α-helical regions of ∼40-80 residues. Notably, these segmentations are evolutionarily conserved between uni- and multicellular eukaryotes. Strikingly, the boundaries of these segments match our previously reported mapping and crystal data, which collectively identified two "cognate" segments of Nup54, each interacting with cognate segments, one in Nup58 and the other one in Nup62. Because Nup54 and Nup58 cognate segments form crystallographic hetero- or homo-oligomers, we proposed that these oligomers associate into inter-convertible "mid-plane" rings: a single large ring (40-50 nm diameter, consisting of eight hetero-dodecamers) or three small rings (10-20 nm diameter, each comprising eight homo-tetramers). Each "ring cycle" would recapitulate "dilation" and "constriction" of the nuclear pore complex's central transport channel. As for the Nup54·Nup62 interactome, it forms a 1:2 triple helix ("finger"), multiples of which project alternately up and down from mid-plane ring(s). Collectively, our previous crystal data suggested a copy number of 128, 64, and 32 for Nup62, Nup54, and Nup58, respectively, that is, a 4:2:1 stoichiometry. Here, we carried out solution analysis utilizing the entire ordered regions of Nup62, Nup54, and Nup58, and demonstrate that they form a dynamic "triple complex" that is heterogeneously formed from our previously characterized Nup54·Nup58 and Nup54·Nup62 interactomes. These data are consistent both with our crystal structure-deduced copy numbers and stoichiometries and also with our ring cycle model for structure and dynamics of the nuclear pore channel.
Keywords: FG nucleoporins; Light scattering; Nuclear Pore Complex; Ring cycle hypothesis; Transport channel; biophysics; cell biology; nuclear pore; nuclear transport; protein assembly.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
FIGURE 1.
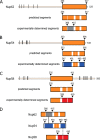
Evolutionarily conserved segmentation of ordered regions of channel nucleoporins, Nup62, Nup54, and Nup58. A–C, top panels, full-length Nup62, Nup54, and Nup58 from rat (R. norvegicus) with boundaries of each predicted ordered region (orange-colored box), disordered region (thin line), and the location of FG repeats (black bars) indicated. Middle panels, predicted α-helical segments (orange) and connecting loops (white). Bottom panels, ordered segments from previously determined crystal structures, color-coded as follows: gray for Nup62, blue for Nup54, and red for the two adjacent segments of Nup58 involved in forming a helix-loop-helix hairpin (11, 16). D, color-coded segments of channel nups that crystallized as hetero-interactomes (Nup54·Nup62) (black line) (9), and as both homo-interactomes and hetero-interactomes (Nup54·Nup58, Nup54 homo-tetramer and Nup58 homo-tetramer) (9, 11, 16). For relevant primary structures of R. norvegicus and Saccharomyces cerevisiae nups, see Fig. 2.
FIGURE 2.
Segmentation topology of each ordered region of the three channel nucleoporins is evolutionarily conserved. A–C, sequence and predicted secondary structure comparisons of ordered region of Nup62/Nsp1 (A); Nup54/Nup57 (B); and Nup58/Nup49 (C) of R. norvegicus/S. cerevisiae, respectively, are shown. Consensus helical regions, as predicted by PROMALS3D, are labeled with h below the sequence alignment. Starting and ending residues of the α-helical regions used in the alignments are numbered. Orange bar highlights the residues of the regions for which x-ray structures are known from R. norvegicus (9, 11, 16).
FIGURE 3.
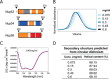
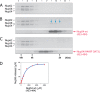
Size-exclusion chromatography elution profiles and circular dichroism of triple complex assembled from the ordered regions of Nup62, Nup54, and Nup58. A, boundaries for each ordered region of channel nups used for the triple complex formation color-coded as Fig. 1_D. B_, elution profiles at increasing concentrations of the triple complex in size-exclusion chromatography, monitored by the absorbance at 280 nm. C, circular dichroism wavelength scan of molar ellipticity of the triple complex acquired at 0.45 mg/ml. Negative ellipticity minima at 208 and 222 nm indicate that the triple complex is primarily α-helical. deg, degrees. D, α-helical contents of triple complexes at increasing concentrations (Conc.) determined by CD spectroscopy.
FIGURE 4.
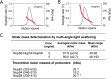
Analysis of triple complex by size-exclusion chromatography coupled to MALS. A and B, Rayleigh ratio (blue line) and molar masses (red line) were determined at two concentrations (A, 2 mg/ml; B, 5 mg/ml) across the elution peak. MW, molar mass. C, summary of averaged molar masses and of mass ranges for the triple complex at two concentrations (Conc.), and theoretical molar masses of the three nucleoporins; asterisk denotes the mass including the peptide sequence (GSHM) from the vector that precedes the actual Nup58 sequence.
FIGURE 5.
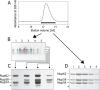
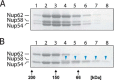
Heterogeneity of triple complex detected by multidimensional analysis. A, fractions across the size-exclusion chromatography peak (horizontal bar) were used for electrophoresis. B, native PAGE. “Iso-electrophoretic” bands were cut separately (marked by red asterisks). C, proteins from these different bands were separated by SDS-PAGE. D, fractions across the size-exclusion chromatography peak (A) analyzed by SDS-PAGE directly.
FIGURE 6.
The triple complex is partially disassembled by non-ionic detergent. A and B, the triple complex (5 mg/ml) was subjected to size-exclusion chromatography in the absence (A) or presence of 65 m
m
_n_-octyl-β-
d
-glucopyranoside (B) followed by SDS-PAGE of column fractions. Blue arrowheads in B indicate that Nup58 was dissociated from the triple complex; black arrows indicate elution positions of molecular weight markers.
FIGURE 7.
The Nup58-cognate segment of Nup54 is solely responsible for tying the ordered region of Nup58 to the triple complex. A–C, triple complex (1.5 mg/ml) was incubated in the absence (A) or in the presence of Nup54 (453–494), using either wild type (0.6 mg/ml) (B) or mutant H469F/Q473L (0.6 mg/ml) (C), followed by size-exclusion chromatography of the incubated sample and subsequent SDS-PAGE of column fractions. Note, that Nup58 (blue arrows in B) is partially removed from the triple complex by wt Nup54 (453–494) (B), but not by mutant Nup54. D, in a matrix depletion assay, where triple complex is immobilized via a Nup62-attached MBP tag, increasing concentrations of wt Nup54 (453–494) led to increasing Nup58 removal.
FIGURE 8.
Protection from limited proteolysis of the triple complex in solution matches regions of cognate segments of previously determined structures. The color gradient of each bar represents the frequency of a fragment observed in mass spectrometry after limited proteolysis: white represents the least protected region, whereas the most protected regions are depicted by dark gray (Nup62), dark blue (Nup54), and dark red (Nup58). Residues of the regions for which x-ray structures are known are colored as orange.
FIGURE 9.
Formation of triple complex requires cognate segments of Nup58 and Nup62. His6-tagged Nup58, representing its full-length ordered region (A) or its C-terminal segment (B) (cognate for Nup54), pull down a triple complex, when co-expressed with Nup54 and Nup62, as assessed by SDS-PAGE (D and E, respectively). In contrast, His6-tagged Nup58 containing only the N-terminal segment of the ordered region of Nup58 is unable to efficiently pull down a triple complex (C and F). Using His6-tagged Nup58 (representing its ordered region) for affinity pulldown and testing truncated forms of the ordered region of Nup62, only the N-terminal segment of Nup62 (cognate to the N-terminal segment of Nup54) (G) was incorporated into a triple complex (I), whereas the C-terminal segment of Nup62 (H) was not (J).
FIGURE 10.
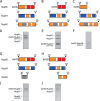
Piecing together crystal structures of channel nucleoporin segments into a model for a nuclear pore channel. A and B, schematic representation of inter-convertible dilated (A) and constricted (B) channel states. The path of segments of ordered regions of channel nups, Nup62 (gray), Nup54 (blue), and Nup58 (red), through the channel's two principal structural elements, mid-plane ring and attached fingers, is shown. The latter are projecting to nucleoplasm and cytoplasm. C and D, a single module (out of eight) is represented each for the dilated (C) or constricted (D) states of the channel. The “linker region” of Nup54 is indicated either by curved lines (A and B) or by dotted lines (C and D).
Similar articles
- Ring cycle for dilating and constricting the nuclear pore.
Solmaz SR, Blobel G, Melcák I. Solmaz SR, et al. Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):5858-63. doi: 10.1073/pnas.1302655110. Epub 2013 Mar 11. Proc Natl Acad Sci U S A. 2013. PMID: 23479651 Free PMC article. - The Nup62 Coiled-Coil Motif Provides Plasticity for Triple-Helix Bundle Formation.
Dewangan PS, Sonawane PJ, Chouksey AR, Chauhan R. Dewangan PS, et al. Biochemistry. 2017 Jun 6;56(22):2803-2811. doi: 10.1021/acs.biochem.6b01050. Epub 2017 Apr 26. Biochemistry. 2017. PMID: 28406021 - The stoichiometry of the nucleoporin 62 subcomplex of the nuclear pore in solution.
Ulrich A, Partridge JR, Schwartz TU. Ulrich A, et al. Mol Biol Cell. 2014 May;25(9):1484-92. doi: 10.1091/mbc.E13-12-0745. Epub 2014 Feb 26. Mol Biol Cell. 2014. PMID: 24574455 Free PMC article. - Structural analysis of the nuclear pore complex by integrated approaches.
Elad N, Maimon T, Frenkiel-Krispin D, Lim RY, Medalia O. Elad N, et al. Curr Opin Struct Biol. 2009 Apr;19(2):226-32. doi: 10.1016/j.sbi.2009.02.009. Epub 2009 Mar 25. Curr Opin Struct Biol. 2009. PMID: 19327984 Review. - Structure and Assembly of the Nuclear Pore Complex.
Hampoelz B, Andres-Pons A, Kastritis P, Beck M. Hampoelz B, et al. Annu Rev Biophys. 2019 May 6;48:515-536. doi: 10.1146/annurev-biophys-052118-115308. Epub 2019 Apr 3. Annu Rev Biophys. 2019. PMID: 30943044 Review.
Cited by
- Molecular and structural analysis of central transport channel in complex with Nup93 of nuclear pore complex.
Sonawane PJ, S Dewangan P, Madheshiya PK, Chopra K, Kumar M, Niranjan S, Ansari MY, Singh J, Bawaria S, Banerjee M, Chauhan R. Sonawane PJ, et al. Protein Sci. 2020 Dec;29(12):2510-2527. doi: 10.1002/pro.3983. Epub 2020 Nov 16. Protein Sci. 2020. PMID: 33085133 Free PMC article. - Conformation of the nuclear pore in living cells is modulated by transport state.
Pulupa J, Prior H, Johnson DS, Simon SM. Pulupa J, et al. Elife. 2020 Dec 21;9:e60654. doi: 10.7554/eLife.60654. Elife. 2020. PMID: 33346731 Free PMC article. - Insights into the role of Nup62 and Nup93 in assembling cytoplasmic ring and central transport channel of the nuclear pore complex.
Madheshiya PK, Shukla E, Singh J, Bawaria S, Ansari MY, Chauhan R. Madheshiya PK, et al. Mol Biol Cell. 2022 Dec 1;33(14):ar139. doi: 10.1091/mbc.E22-01-0027. Epub 2022 Oct 12. Mol Biol Cell. 2022. PMID: 36222862 Free PMC article. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review. - Architecture of the fungal nuclear pore inner ring complex.
Stuwe T, Bley CJ, Thierbach K, Petrovic S, Schilbach S, Mayo DJ, Perriches T, Rundlet EJ, Jeon YE, Collins LN, Huber FM, Lin DH, Paduch M, Koide A, Lu V, Fischer J, Hurt E, Koide S, Kossiakoff AA, Hoelz A. Stuwe T, et al. Science. 2015 Oct 2;350(6256):56-64. doi: 10.1126/science.aac9176. Epub 2015 Aug 27. Science. 2015. PMID: 26316600 Free PMC article.
References
- Hoelz A., Debler E. W., Blobel G. (2011) The structure of the nuclear pore complex. Annu. Rev. Biochem. 80, 613–643 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials