A large multiethnic genome-wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences - PubMed (original) (raw)
doi: 10.1158/2159-8290.CD-15-0315. Epub 2015 Jun 1.
Stephen K Van Den Eeden 2, Lori C Sakoda 3, Eric Jorgenson 3, Laurel A Habel 3, Rebecca E Graff 4, Michael N Passarelli 4, Clinton L Cario 4, Nima C Emami 4, Chun R Chao 5, Nirupa R Ghai 5, Jun Shan 3, Dilrini K Ranatunga 3, Charles P Quesenberry 3, David Aaronson 6, Joseph Presti 6, Zhaoming Wang 7, Sonja I Berndt 7, Stephen J Chanock 7, Shannon K McDonnell 8, Amy J French 9, Daniel J Schaid 8, Stephen N Thibodeau 9, Qiyuan Li 10, Matthew L Freedman 11, Kathryn L Penney 12, Lorelei A Mucci 12, Christopher A Haiman 13, Brian E Henderson 13, Daniela Seminara 14, Mark N Kvale 15, Pui-Yan Kwok 15, Catherine Schaefer 3, Neil Risch 16, John S Witte 17
Affiliations
- PMID: 26034056
- PMCID: PMC4527942
- DOI: 10.1158/2159-8290.CD-15-0315
A large multiethnic genome-wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences
Thomas J Hoffmann et al. Cancer Discov. 2015 Aug.
Abstract
A genome-wide association study (GWAS) of prostate cancer in Kaiser Permanente health plan members (7,783 cases, 38,595 controls; 80.3% non-Hispanic white, 4.9% African-American, 7.0% East Asian, and 7.8% Latino) revealed a new independent risk indel rs4646284 at the previously identified locus 6q25.3 that replicated in PEGASUS (N = 7,539) and the Multiethnic Cohort (N = 4,679) with an overall P = 1.0 × 10(-19) (OR, 1.18). Across the 6q25.3 locus, rs4646284 exhibited the strongest association with expression of SLC22A1 (P = 1.3 × 10(-23)) and SLC22A3 (P = 3.2 × 10(-52)). At the known 19q13.33 locus, rs2659124 (P = 1.3 × 10(-13); OR, 1.18) nominally replicated in PEGASUS. A risk score of 105 known risk SNPs was strongly associated with prostate cancer (P < 1.0 × 10(-8)). Comparing the highest to lowest risk score deciles, the OR was 6.22 for non-Hispanic whites, 5.82 for Latinos, 3.77 for African-Americans, and 3.38 for East Asians. In non-Hispanic whites, the 105 risk SNPs explained approximately 7.6% of disease heritability. The entire GWAS array explained approximately 33.4% of heritability, with a 4.3-fold enrichment within DNaseI hypersensitivity sites (P = 0.004).
Significance: Taken together, our findings of independent risk variants, ethnic variation in existing SNP replication, and remaining unexplained heritability have important implications for further clarifying the genetic risk of prostate cancer. Our findings also suggest that there may be much promise in evaluating understudied variation, such as indels and ethnically diverse populations.
©2015 American Association for Cancer Research.
Conflict of interest statement
Conflict of Interest: No potential conflicts of interest were disclosed.
Figures
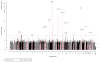
Figure 1
Results from a genome-wide association study (GWAS) of prostate cancer in Kaiser Permanente population (8,399 cases and 38,745 controls), highlighting key chromosomal regions. P-values are for variant associations with prostate cancer from trans-ethnic fixed-effects meta-analysis of four race/ethnicity GWAS (non-Hispanic white, Latino, African-American, East Asian), each adjusted for age and ancestry principal components. Horizontal dashed lines indicated genome-wide statistical significance (p<5×10−8), Bonferroni-corrected significance for 105 know prostate cancer risk variants (p<0.05/105), and nominal significance (p<0.05). Red points denote our findings for the 105 known risk SNPs, and pink indicate results for SNPs within a 0.5Mb window around these SNPs. The new 6q25.3 indel rs4646284 is colored magenta, and the 19q13.33 prostate cancer or PSA SNP rs2659124 is noted in green. Those loci containing previously-reported variants that we replicated at genome-wide significance are noted in black text. Loci with variants that were novel in in the KP GWAS but failed to replicate are noted in gray text.
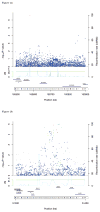
Figure 2
Regional fixed-effects meta-analysis plots from GWAS in Kaiser Permanente population of the two risk variants that replicated: (a) 6q25.3 (rs4646284), (b) 19q13.33 (rs2659124). The color code for the points represents the r2 of each SNP with the risk variant (ranges defined in the legend). The dotted vertical lines pass through the risk variants and the other significant SNPs in the region, on which analyses were conditioned.
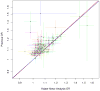
Figure 3
Comparison of variant associations from previous reports and results from the KP GWAS meta-analysis for 105 known prostate cancer risk SNPs. Plotted values are the log odds ratios from the previous and current studies. Error lines denote the 95% CIs for the respective studies, colored by chromosome. Blue line is the diagonal and red is regression fit through the points, showing extremely high correlation between the previous and new odds ratios. The SNP rs116041037 appears to be an African-American specific SNP and is an outlier so we left it off of the plot to more easily view the other points; the SNP had a previous OR=2.45 (95% CI=1.65–3.62), and KP OR=2.67 (1.96, 3.64).
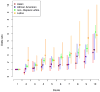
Figure 4
Impact of increasing deciles of risk profile scores based on 105 known risk SNPs across different race/ethnicity groups. Risk scores generated by combining the 105 SNPs into a single score, applying the logs odds ratio estimated by previous studies to our KP study population genotype data. Odds ratios for effect of risk profile scores on prostate cancers calculated within each decile of the scores, using the lowest decile of risk profile scores as the referent category. The non-Hispanic white and Latino groups had substantially higher ORs than the African-Americans, and the East Asians always had the lowest ORs.
Similar articles
- Evaluating genetic risk for prostate cancer among Japanese and Latinos.
Cheng I, Chen GK, Nakagawa H, He J, Wan P, Laurie CC, Shen J, Sheng X, Pooler LC, Crenshaw AT, Mirel DB, Takahashi A, Kubo M, Nakamura Y, Al Olama AA, Benlloch S, Donovan JL, Guy M, Hamdy FC, Kote-Jarai Z, Neal DE, Wilkens LR, Monroe KR, Stram DO, Muir K, Eeles RA, Easton DF, Kolonel LN, Henderson BE, Le Marchand L, Haiman CA. Cheng I, et al. Cancer Epidemiol Biomarkers Prev. 2012 Nov;21(11):2048-58. doi: 10.1158/1055-9965.EPI-12-0598. Epub 2012 Aug 24. Cancer Epidemiol Biomarkers Prev. 2012. PMID: 22923026 Free PMC article. - Multi-ethnic transcriptome-wide association study of prostate cancer.
Fiorica PN, Schubert R, Morris JD, Abdul Sami M, Wheeler HE. Fiorica PN, et al. PLoS One. 2020 Sep 28;15(9):e0236209. doi: 10.1371/journal.pone.0236209. eCollection 2020. PLoS One. 2020. PMID: 32986714 Free PMC article. - Generalizability of associations from prostate cancer genome-wide association studies in multiple populations.
Waters KM, Le Marchand L, Kolonel LN, Monroe KR, Stram DO, Henderson BE, Haiman CA. Waters KM, et al. Cancer Epidemiol Biomarkers Prev. 2009 Apr;18(4):1285-9. doi: 10.1158/1055-9965.EPI-08-1142. Epub 2009 Mar 24. Cancer Epidemiol Biomarkers Prev. 2009. PMID: 19318432 Free PMC article. - Identification of novel common breast cancer risk variants at the 6q25 locus among Latinas.
Hoffman J, Fejerman L, Hu D, Huntsman S, Li M, John EM, Torres-Mejia G, Kushi L, Ding YC, Weitzel J, Neuhausen SL, Lott P; COLUMBUS Consortium; Echeverry M, Carvajal-Carmona L, Burchard E, Eng C, Long J, Zheng W, Olopade O, Huo D, Haiman C, Ziv E. Hoffman J, et al. Breast Cancer Res. 2019 Jan 14;21(1):3. doi: 10.1186/s13058-018-1085-9. Breast Cancer Res. 2019. PMID: 30642363 Free PMC article. - A systematic review of replication studies of prostate cancer susceptibility genetic variants in high-risk men originally identified from genome-wide association studies.
Ishak MB, Giri VN. Ishak MB, et al. Cancer Epidemiol Biomarkers Prev. 2011 Aug;20(8):1599-610. doi: 10.1158/1055-9965.EPI-11-0312. Epub 2011 Jun 29. Cancer Epidemiol Biomarkers Prev. 2011. PMID: 21715604 Review.
Cited by
- Pleiotropic Locus 15q24.1 Reveals a Gender-Specific Association with Neovascular but Not Atrophic Age-Related Macular Degeneration (AMD).
Kiel C, Strunz T, International Amd Genomics Consortium Project Manager Susan Blanton Iamdgc, Grassmann F, Weber BHF. Kiel C, et al. Cells. 2020 Oct 8;9(10):2257. doi: 10.3390/cells9102257. Cells. 2020. PMID: 33050031 Free PMC article. - Phytochemicals in Prostate Cancer: From Bioactive Molecules to Upcoming Therapeutic Agents.
Salehi B, Fokou PVT, Yamthe LRT, Tali BT, Adetunji CO, Rahavian A, Mudau FN, Martorell M, Setzer WN, Rodrigues CF, Martins N, Cho WC, Sharifi-Rad J. Salehi B, et al. Nutrients. 2019 Jun 29;11(7):1483. doi: 10.3390/nu11071483. Nutrients. 2019. PMID: 31261861 Free PMC article. Review. - Divergent effects of vitamins K1 and K2 on triple negative breast cancer cells.
Beaudin S, Kokabee L, Welsh J. Beaudin S, et al. Oncotarget. 2019 Mar 19;10(23):2292-2305. doi: 10.18632/oncotarget.26765. eCollection 2019 Mar 19. Oncotarget. 2019. PMID: 31040920 Free PMC article. - Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study.
Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Mertens J, Weisner C, Choquet H. Jorgenson E, et al. Mol Psychiatry. 2017 Sep;22(9):1359-1367. doi: 10.1038/mp.2017.101. Epub 2017 May 9. Mol Psychiatry. 2017. PMID: 28485404 Free PMC article. - African American Prostate Cancer Displays Quantitatively Distinct Vitamin D Receptor Cistrome-transcriptome Relationships Regulated by BAZ1A.
Siddappa M, Hussain S, Wani SA, White J, Tang H, Gray JS, Jafari H, Wu HC, Long MD, Elhussin I, Karanam B, Wang H, Morgan R, Hardiman G, Adelani IB, Rotimi SO, Murphy AR, Nonn L, Davis MB, Kittles RA, Hughes Halbert C, Sucheston-Campbell LE, Yates C, Campbell MJ. Siddappa M, et al. Cancer Res Commun. 2023 Apr 18;3(4):621-639. doi: 10.1158/2767-9764.CRC-22-0389. eCollection 2023 Apr. Cancer Res Commun. 2023. PMID: 37082578 Free PMC article.
References
- Hayes RB, Liff JM, Pottern LM, Greenberg RS, Schoenberg JB, Schwartz AG, et al. Prostate cancer risk in U.S. blacks and whites with a family history of cancer. Int J Cancer J Int Cancer. 1995;60:361–4. - PubMed
- Whittemore AS, Wu AH, Kolonel LN, John EM, Gallagher RP, Howe GR, et al. Family history and prostate cancer risk in black, white, and Asian men in the United States and Canada. Am J Epidemiol. 1995;141:732–40. - PubMed
- Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;13(Spec No 1):R103–21. - PubMed
- Baker SG, Lichtenstein P, Kaprio J, Holm N. Genetic susceptibility to prostate, breast, and colorectal cancer among Nordic twins. Biometrics. 2005;61:55–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA54281/CA/NCI NIH HHS/United States
- T32 EB009383/EB/NIBIB NIH HHS/United States
- P30 CA082103/CA/NCI NIH HHS/United States
- CA148085/CA/NCI NIH HHS/United States
- P30 CA015083/CA/NCI NIH HHS/United States
- R01 CA088164/CA/NCI NIH HHS/United States
- R01 CA136578/CA/NCI NIH HHS/United States
- R37 CA054281/CA/NCI NIH HHS/United States
- R01 CA141298/CA/NCI NIH HHS/United States
- RC2 AG036607/AG/NIA NIH HHS/United States
- R01 CA151254/CA/NCI NIH HHS/United States
- RC2 CA148085/CA/NCI NIH HHS/United States
- CA127298/CA/NCI NIH HHS/United States
- CA136578/CA/NCI NIH HHS/United States
- CA112355/CA/NCI NIH HHS/United States
- CA1326792/CA/NCI NIH HHS/United States
- CA131945/CA/NCI NIH HHS/United States
- T32 GM067547/GM/NIGMS NIH HHS/United States
- CA088164/CA/NCI NIH HHS/United States
- T32 CA112355/CA/NCI NIH HHS/United States
- U01 CA127298/CA/NCI NIH HHS/United States
- CA141298/CA/NCI NIH HHS/United States
- R01 CA157881/CA/NCI NIH HHS/United States
- R25 CA112355/CA/NCI NIH HHS/United States
- R01 GM107427/GM/NIGMS NIH HHS/United States
- HG004726/HG/NHGRI NIH HHS/United States
- AG046616/AG/NIA NIH HHS/United States
- CA63464/CA/NCI NIH HHS/United States
- K01 DC013300/DC/NIDCD NIH HHS/United States
- DC013300/DC/NIDCD NIH HHS/United States
- R01 CA131945/CA/NCI NIH HHS/United States
- CA151254/CA/NCI NIH HHS/United States
- R01 CA063464/CA/NCI NIH HHS/United States
- U01 CA063464/CA/NCI NIH HHS/United States
- CA157881/CA/NCI NIH HHS/United States
- R01 CA054281/CA/NCI NIH HHS/United States
- R21 AG046616/AG/NIA NIH HHS/United States
- U01 HG004726/HG/NHGRI NIH HHS/United States