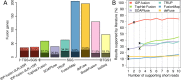Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing - PubMed (original) (raw)
Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing
Jason L Weirather et al. Nucleic Acids Res. 2015.
Abstract
We developed an innovative hybrid sequencing approach, IDP-fusion, to detect fusion genes, determine fusion sites and identify and quantify fusion isoforms. IDP-fusion is the first method to study gene fusion events by integrating Third Generation Sequencing long reads and Second Generation Sequencing short reads. We applied IDP-fusion to PacBio data and Illumina data from the MCF-7 breast cancer cells. Compared with the existing tools, IDP-fusion detects fusion genes at higher precision and a very low false positive rate. The results show that IDP-fusion will be useful for unraveling the complexity of multiple fusion splices and fusion isoforms within tumorigenesis-relevant fusion genes.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
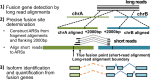
Figure 1.
Flowchart of IDP-fusion. IDP-fusion contains three main steps: (1) detect fusion genes by genome-wide alignments of long reads (e.g. using GMAP); (2) determine precise fusion sites by alignment of short reads to Artificial Reference Sequences (ARSs) (e.g. using STAR); (3) identify and quantify significantly expressed isoforms, including fusion isoforms, from fusion genes.
Figure 2.
Isoform candidate constructions from fusion genes. Isoform candidate constructions contain three structural units: 5′/3′ ends, non-redundant splice linkages (including fusion splice linkages) and splices. All possible splice combinations are included until reaching the 5′ end and 3′ end. Both regular isoforms and fusion isoforms are constructed.
Figure 3.
Precisions of fusion gene detections by IDP-fusion, six SGS-only methods and a TGS-only method. (A) The total numbers of fusion genes detected from MCF-7 cells by IDP-fusion, six SGS-only methods and Iso-Seq (TGS-only method) and the corresponding numbers of the gold standard fusion genes are shown by stacked bars. The precisions are also shown as the rates of the gold standard fusion genes detected. Two modes of IDP-fusion, using either ≥2 supporting long reads (LR) and ≥1 supporting short read (SR) or ≥2 supporting long reads and ≥2 supporting short reads (default settings) are compared with six SGS-only methods that were run under their default settings and Iso-Seq method requiring ≥5 full-length long reads. (B) The precision of fusion gene detection by IDP-fusion is higher than SGS-only methods, regardless of increase of the minimum numbers of supporting short reads. The precision of the default setting for each method is labeled by ‘x’. Note that BreakFusion is not shown because the software does not output the number of supporting reads for each fusion site.
Figure 4.
Precise fusion site determination of the fusion gene AIB1–chr1:107078407. (A) A fusion between AIB1 and an unannotated region of chromosome 1 was detected by six long reads (blue blocks), but the long read alignment ends are not in agreement on the precise fusion site. The alignment ends of the long read fragments span 37 and 65 bp at two sides, respectively. IDP-fusion aligned 31 short reads (black blocks) to the fusion site precisely at chr20:46130763–chr1:107078407. In particular, chr20:46130763 is the 3′ end of the first exon of AIB1 and the canonical splicing signal is found in this fusion site. (B) The fusion site chr20:46130763–chr1:107078407 was PCR validated from MCF-7 cDNA, but not healthy breast cDNA (BC) or a genomic control (GC).
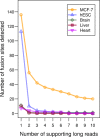
Figure 5.
The numbers of fusion sites determined from MCF-7 cells and four normal samples. Compared with the breast cancer cells MCF-7, four negative controls (human embryonic stem cell line/hESC, and human brain, liver and heart) are expected to have negligible gene fusion events. As increasing the requirement of supporting long reads, the numbers of fusion sites decreases. The dramatic drop occurs at the requirement of ≥2 supporting long reads. The fusion sites determined from the negative controls are negligible.
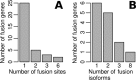
Figure 6.
The distribution of fusion sites and fusion isoform counts in fusion genes of MCF-7 cells. (A) Single fusion sites were determined from 25 fusion genes and multiple fusion sites in the other 10 fusion genes. In particular, six fusion sites were determined in both fusion genes AIB1–chr1:107078407 and BCAS4–BCAS3. (B) Significantly expressed fusion isoforms (RPKM > 10) were identified from 14 fusion genes. Eight significantly expressed fusion isoforms were identified from BCAS4-BCAS3 (Figure 7A).
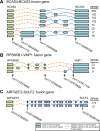
Figure 7.
Illustration of fusion isoforms occurring through alternative fusion splices or alternative regular splices. (A) Eight significantly expressed fusion isoforms were identified and quantified in the fusion gene BCAS4-BCAS3, sharing three fusion splices. Besides alternative fusion splices, the regular alternative splices within BCAS3 also contribute to the fusion isoform generation. (B) Three significantly expressed fusion isoforms were identified and quantified in the fusion gene RPS6KB1–VMP1, sharing two fusion splices. Besides alternative fusion splices, the regular alternative splices within both RPS6KB1 and VMP1 also contribute to the fusion isoform generation. (C) Two significantly expressed fusion isoforms were identified and quantified in the fusion gene ARFGEF2–SULF2, sharing only one fusion splice. The diversity of fusion isoform expression is driven by the regular alternative splicing of SULF2.
Figure 8.
IDP-fusion detected and annotated fusion genes with novel gene and novel exons involved. (A) IDP-fusion detected the fusion gene between AIB1 and an unannotated region in chromosome 1. Eight fusion isoforms were estimated at the modest abundance (RPKM > 1) and contain seven novel exons (blue) annotated by IDP-fusion. (B) A selection of fusion long read alignments (bright blue) contributing structural information to isoform reconstruction is displayed in line with four reference annotations libraries: UCSC, RefSeq, GENCODE and Ensembl. The long read fragment alignments to AIB1 gene locus are shown at the left and the ones to the novel gene locus at the right. The long read alignments annotated a novel gene with seven exons not reported by the reference annotation libraries. (C) IDP-fusion detected the fusion gene UNK–ABCA5 and two corresponding fusion isoforms with RPKM > 1. This fusion gene involves two novel exons (in red box) upstream of ABCA5. (D) A selection of fusion long read alignments (bright blue) contributing structural information to isoform reconstruction is displayed in line with four reference annotations libraries: UCSC, RefSeq, GENCODE and Ensembl. The long read fragment alignments to UNK gene locus are shown at the left and the ones to ABCA5 gene locus at the right. The long read alignments detected two novel exons (in red box), while they are not reported by the reference annotation libraries. Note that ABCA5 is transcribed from the reverse strand. To show the fusion gene in the correct order, the browser figure of the ABCA5 gene locus is flipped. Please refer to Figure 7 for a description of figure elements.
References
- Edwards P.A. Fusion genes and chromosome translocations in the common epithelial cancers. J. Pathol. 2010;220:244–254. -PubMed
- Vega F., Medeiros L.J. Chromosomal translocations involved in non-Hodgkin lymphomas. Arch. Pathol. Lab. Med. 2003;127:1148–1160. -PubMed
- Nowell P., Hungerford D.A. A minute chromosome in human chronic granulocytic leukemia [abstract] Science. 1960;132:1488–1501.
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM109836/GM/NIGMS NIH HHS/United States
- R01 HG007834/HG/NHGRI NIH HHS/United States
- R01GM109836/GM/NIGMS NIH HHS/United States
- R01HG007834/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources