High-Resolution Analysis by Whole-Genome Sequencing of an International Lineage (Sequence Type 111) of Pseudomonas aeruginosa Associated with Metallo-Carbapenemases in the United Kingdom - PubMed (original) (raw)
. 2015 Aug;53(8):2622-31.
doi: 10.1128/JCM.00505-15. Epub 2015 Jun 3.
Laura Wright 2, Anthony Underwood 3, Adam A Witney 4, Yuen-Ting Chan 5, Ali Al-Shahib 3, Catherine Arnold 5, Michel Doumith 2, Bharat Patel 6, Timothy D Planche 7, Jonathan Green 3, Richard Holliman 8, Neil Woodford 2
Affiliations
- PMID: 26041902
- PMCID: PMC4508460
- DOI: 10.1128/JCM.00505-15
High-Resolution Analysis by Whole-Genome Sequencing of an International Lineage (Sequence Type 111) of Pseudomonas aeruginosa Associated with Metallo-Carbapenemases in the United Kingdom
Jane F Turton et al. J Clin Microbiol. 2015 Aug.
Abstract
Whole-genome sequencing (WGS) was carried out on 87 isolates of sequence type 111 (ST-111) of Pseudomonas aeruginosa collected between 2005 and 2014 from 65 patients and 12 environmental isolates from 24 hospital laboratories across the United Kingdom on an Illumina HiSeq instrument. Most isolates (73) carried VIM-2, but others carried IMP-1 or IMP-13 (5) or NDM-1 (1); one isolate had VIM-2 and IMP-18, and 7 carried no metallo-beta-lactamase (MBL) gene. Single nucleotide polymorphism analysis divided the isolates into distinct clusters; the NDM-1 isolate was an outlier, and the IMP isolates and 6/7 MBL-negative isolates clustered separately from the main set of 73 VIM-2 isolates. Within the VIM-2 set, there were at least 3 distinct clusters, including a tightly clustered set of isolates from 3 hospital laboratories consistent with an outbreak from a single introduction that was quickly brought under control and a much broader set dominated by isolates from a long-running outbreak in a London hospital likely seeded from an environmental source, requiring different control measures; isolates from 7 other hospital laboratories in London and southeast England were also included. Bayesian evolutionary analysis indicated that all the isolates shared a common ancestor dating back ∼50 years (1960s), with the main VIM-2 set separating approximately 20 to 30 years ago. Accessory gene profiling revealed blocks of genes associated with particular clusters, with some having high similarity (≥95%) to bacteriophage genes. WGS of widely found international lineages such as ST-111 provides the necessary resolution to inform epidemiological investigations and intervention policies.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
FIG 1
Charts showing numbers of patients (shown by metallo-beta-lactamase status in panel a and additionally with number of hospital laboratories [requestors] in panel b) from which representatives of ST-111 were received between 2005 and April 2014. Patients are included only once (by year when the first isolate was received), but requestors are included for every year that they submitted isolates from new patients. “Not tested” applies to isolates subjected only to typing that are no longer available in our archives; all but one was from hospital London_17, and it is assumed that they are highly likely to be VIM positive.
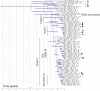
FIG 2
Maximum-likelihood tree estimation using single nucleotide polymorphism (SNP) analysis of representatives of ST-111 from United Kingdom hospitals collected between 2005 and 2014. Isolates are labeled by patient (P1 to P64 and S1 to S4), hospital, metallo-beta-lactamase gene (where present), and date (month.year) of collection. Environmental isolates are labeled E1 to E12, followed by this information. Two isolates from patient P8, separated by 4 years, are marked with stars; other patient pairs are marked with triangles, circles, squares, diamonds, or hexagons. Clusters and subclusters described in the text are marked. Bootstrap values are given for the clusters described.
FIG 3
Bayesian evolutionary analysis sampling trees (BEAST) analysis of representatives of ST-111 from United Kingdom hospitals collected between 2004 and 2014 calculated using a relaxed molecular clock. Isolates and labeling are as in Fig. 2. The horizontal position of isolates on the tree is according to the date that they were isolated; calculated times for putative common ancestors (at the nodes) can be read from the scale (in years). Time zero is that of the most recent isolate (April 2014). Horizontal bars indicate degrees of confidence.
FIG 4
Presence/absence of accessory genes in representatives of ST-111 from United Kingdom hospitals collected between 2005 and 2014 obtained from an iterative-BLAST approach. Vertical black bars represent presence of accessory genes displayed against a phylogenetic tree. A cluster of genes distinguishing the cluster 3 isolates is marked with the red bar and box. A full list of the genes presented is available (see Table S3 in the supplemental material).
FIG 5
Snapshot of accessory gene comparison for some genes of a subset of 24 isolates from various clusters from CD-HIT analysis. Dark squares indicate isolates with particular sets of genes, the number of genes in the set being indicated in the bottom row, e.g., the starred set indicates 44 genes exclusive to clusters 1 and 2 isolates among the set.
Similar articles
- Dissemination of IMP-6 metallo-β-lactamase-producing Pseudomonas aeruginosa sequence type 235 in Korea.
Seok Y, Bae IK, Jeong SH, Kim SH, Lee H, Lee K. Seok Y, et al. J Antimicrob Chemother. 2011 Dec;66(12):2791-6. doi: 10.1093/jac/dkr381. Epub 2011 Sep 19. J Antimicrob Chemother. 2011. PMID: 21933788 - Molecular evolution of metallo-beta-lactamase-producing Pseudomonas aeruginosa in a nosocomial setting of high-level endemicity.
Lagatolla C, Edalucci E, Dolzani L, Riccio ML, De Luca F, Medessi E, Rossolini GM, Tonin EA. Lagatolla C, et al. J Clin Microbiol. 2006 Jul;44(7):2348-53. doi: 10.1128/JCM.00258-06. J Clin Microbiol. 2006. PMID: 16825348 Free PMC article. - Dominance of international 'high-risk clones' among metallo-β-lactamase-producing Pseudomonas aeruginosa in the UK.
Wright LL, Turton JF, Livermore DM, Hopkins KL, Woodford N. Wright LL, et al. J Antimicrob Chemother. 2015 Jan;70(1):103-10. doi: 10.1093/jac/dku339. Epub 2014 Sep 1. J Antimicrob Chemother. 2015. PMID: 25182064 - Metallo-beta-lactamase-producing Pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones.
Juan C, Zamorano L, Mena A, Albertí S, Pérez JL, Oliver A. Juan C, et al. J Antimicrob Chemother. 2010 Mar;65(3):474-8. doi: 10.1093/jac/dkp491. Epub 2010 Jan 12. J Antimicrob Chemother. 2010. PMID: 20071364 - Developing insights into the mechanisms of evolution of bacterial pathogens from whole-genome sequences.
Bryant J, Chewapreecha C, Bentley SD. Bryant J, et al. Future Microbiol. 2012 Nov;7(11):1283-1296. doi: 10.2217/fmb.12.108. Future Microbiol. 2012. PMID: 23075447 Free PMC article. Review.
Cited by
- Diversity and Distribution of Resistance Markers in Pseudomonas aeruginosa International High-Risk Clones.
Kocsis B, Gulyás D, Szabó D. Kocsis B, et al. Microorganisms. 2021 Feb 12;9(2):359. doi: 10.3390/microorganisms9020359. Microorganisms. 2021. PMID: 33673029 Free PMC article. Review. - Past and Present Perspectives on β-Lactamases.
Bush K. Bush K. Antimicrob Agents Chemother. 2018 Sep 24;62(10):e01076-18. doi: 10.1128/AAC.01076-18. Print 2018 Oct. Antimicrob Agents Chemother. 2018. PMID: 30061284 Free PMC article. Review. - Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections.
Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, Benito N, Grau S. Horcajada JP, et al. Clin Microbiol Rev. 2019 Aug 28;32(4):e00031-19. doi: 10.1128/CMR.00031-19. Print 2019 Sep 18. Clin Microbiol Rev. 2019. PMID: 31462403 Free PMC article. Review. - High quality 3C de novo assembly and annotation of a multidrug resistant ST-111 Pseudomonas aeruginosa genome: Benchmark of hybrid and non-hybrid assemblers.
Molina-Mora JA, Campos-Sánchez R, Rodríguez C, Shi L, García F. Molina-Mora JA, et al. Sci Rep. 2020 Jan 29;10(1):1392. doi: 10.1038/s41598-020-58319-6. Sci Rep. 2020. PMID: 31996747 Free PMC article. - Outbreak of Pseudomonas aeruginosa producing VIM carbapenemase in an intensive care unit and its termination by implementation of waterless patient care.
Catho G, Martischang R, Boroli F, Chraïti MN, Martin Y, Koyluk Tomsuk Z, Renzi G, Schrenzel J, Pugin J, Nordmann P, Blanc DS, Harbarth S. Catho G, et al. Crit Care. 2021 Aug 19;25(1):301. doi: 10.1186/s13054-021-03726-y. Crit Care. 2021. PMID: 34412676 Free PMC article.
References
- Mulet X, Cabot G, Ocampo-Sosa AA, Domínguez MA, Zamorano L, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Oliver A; Spanish Network for Research in Infectious Diseases (REIPI). 2013. Biological markers of Pseudomonas aeruginosa epidemic high-risk clones. Antimicrob Agents Chemother 57:5527–5535. doi:10.1128/AAC.01481-13. - DOI - PMC - PubMed
- Snyder LA, Loman NJ, Faraj LA, Levi K, Weinstock G, Boswell TC, Pallen MJ, Ala'Aldeen DA. 2013. Epidemiological investigation of Pseudomonas aeruginosa isolates from a six-year-long hospital outbreak using high-throughput whole genome sequencing. Euro Surveill 18(42): pii=20611 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20611. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous