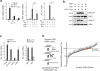PQBP1 Is a Proximal Sensor of the cGAS-Dependent Innate Response to HIV-1 - PubMed (original) (raw)
. 2015 Jun 4;161(6):1293-1305.
doi: 10.1016/j.cell.2015.04.050.
Monika Schneider 1, Janna Seifried # 2, Stephen Soonthornvacharin # 1, Rana E Akleh 1, Kevin C Olivieri 1, Paul D De Jesus 1, Chunhai Ruan 3, Elisa de Castro 4, Pedro A Ruiz 1, David Germanaud 5, Vincent des Portes 6, Adolfo García-Sastre 4 7, Renate König 2 1 8, Sumit K Chanda 1
Affiliations
- PMID: 26046437
- PMCID: PMC4503237
- DOI: 10.1016/j.cell.2015.04.050
PQBP1 Is a Proximal Sensor of the cGAS-Dependent Innate Response to HIV-1
Sunnie M Yoh et al. Cell. 2015.
Abstract
Dendritic cells (DCs) play a critical role in the immune response to viral infection through the facilitation of cell-intrinsic antiviral activity and the activation of adaptive immunity. HIV-1 infection of DCs triggers an IRF3-dependent innate immune response, which requires the activity of cyclic GAMP synthase (cGAS). We report the results of a targeted RNAi screen utilizing primary human monocyte-derived DCs (MDDCs) to identify immune regulators that directly interface with HIV-1-encoded features to initiate this innate response. Polyglutamine binding protein 1 (PQBP1) emerged as a strong candidate through this analysis. We found that PQBP1 directly binds to reverse-transcribed HIV-1 DNA and interacts with cGAS to initiate an IRF3-dependent innate response. MDDCs derived from Renpenning syndrome patients, who harbor mutations in the PQBP1 locus, possess a severely attenuated innate immune response to HIV-1 challenge, underscoring the role of PQBP1 as a proximal innate sensor of a HIV-1 infection.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
Figure 1. A targeted RNAi screen to identify innate immune sensors of HIV-1 infection
(A) MDDCs were infected with HIV/Vpx encoding luciferase for the indicated duration (left). ISG54 mRNA levels were measured after infection in the absence or presence of antiviral drugs, NVP (5 μM) or Ral (5μM) (middle). Viral infectivity was evaluated by assessing viral luciferase activity (right). (B) MDDCs treated as in (A) were assayed for p-IRF3 and p-IKKε levels by western blot. (C) MDDCs were transfected with siRNAs targeting IRF3 and NF-κB (p65), and relative ISG54 induction was measured 12 hr post infection. ‘None’ indicates that the cells were not transfected. (D) A schematic depicting the screening process (left). The average and standard deviation of ISG54 mRNA levels in MDDCs depleted of each putative co-sensor is shown (right). The scramble control activities are depicted in the large, red circles. The horizontal dotted line reflects the 3 standard deviation cut-off from the controls. The p value was determined by one-tailed, unpaired t-test (p≤0.0001). This screen was run in biological duplicates. Except in (D), all data are shown as the average ± SD of biological triplicates. *p < 0.05 as determined by 1 way ANOVA with Tukey's post-test. Panels (A) - (C) represent data from at least three independent experiments. See also Figure S1 and SI Table 1 for additional validation studies of the screening platform.
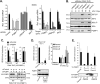
Figure 2. PQBP1 regulates the innate immune response to HIV-1 in primary human DCs and human monocytic cell lines
(A) ISG54 expression was measured in MDDCs transfected with four independent siRNAs targeting PQBP1 and challenged with HIV/Vpx for 8 hr (left). A scrambled siRNA and a siRNA targeting an unrelated innate regulator (LRRFIP1) were utilized as negative controls, and a siRNA targeting IRF3 was employed as a positive control. Expression of each gene was calculated relative to mRNA levels after transfection with the scramble siRNA (right) (* represents a significant [p < 0.05] difference from the scramble). (B) THP-1 were transfected with siRNAs targeting the indicated genes and a scramble control for 48 hr, followed by HIV/Vpx infection; the indicated protein levels were then analyzed. (C) V5-tagged WT-PQBP1-THP-1 (expressing wild type PQBP1) or siR-PQBP1-THP-1 (expressing siRNA-resistant PQBP1) cells were transfected with either a control or PQBP1-targeting siRNA and infected with HIV/Vpx for 8 hr, then the fold induction of ISG54 in comparison to uninfected sample was measured (top panel). V5-tagged PQBP1 protein expression is shown (bottom panel). (D) CRISPR-generated PQBP1 THP-1 cells were PMA-differentiated and infected with HIV/Vpx or stimulated with HT-DNA for 8 hr, then ISG54 induction was measured (top). Loss of PQBP1 protein in cPQBP1 cells was confirmed by western blot (bottom). (E) cPQBP1 clones were stably reconstituted with V5-tagged wild type PQBP1 and infected with HIV/Vpx, and the ISG54 fold induction was compared in all lines (top). Expression of V5-PQBP1 protein in the rescue lines is shown (bottom). Panels from (A) and (B) represent data from at least three independent experiments. Panel (C) – (E) is a representative of two independent experiments, and the data are shown as the mean ± SD of biological triplicates. See also Figure S2.
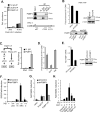
Figure 3. PQBP1 associates with immunogenic HIV-1 DNA
(A) MDDCs were challenged with HIV/Vpx for 3 or 16 hr, followed by formaldehyde cross-linking. The abundance of HIV-1-encoded nucleic acids associating with the PQBP1 IP or the ZC3H3 IP was evaluated using qRT-PCR using primers for the HIV-1 strong stop sequence (left) (eRT primers, Supplementary Data Table 2). The values are relative to the level of nucleic acids that precipitated by normal IgG from the 3 hr infected lysate. Binding specificity of the antibodies was confirmed by western blot of the immunoprecipitants (right). The schematic depicts the HIV-1 genome with the three sets of primers used to measure the amount of HIV-1 nucleic acid. See also Figure S3C. (B) THP-1 cells that stably expressed shRNA targeting luciferase or cGAS were infected with HIV/Vpx for 6 hr, formaldehyde cross-linked, lysed and immunoprecipitated with a PQBP1 antibody. The fold enrichment of HIV-1 DNA (3'LTR) pulled down by the PQBP1-IP over normal IgG-IP is indicated (top). A western blot of cGAS protein in shcGAS cells compared to shLuc control (right). A western blot shows levels of PQBP1 protein immunoprecipitated from both samples (bottom). (C) Immunoprecipitants described in (A) were treated with either DNase or RNaseA/H, as shown in schematic, and HIV-1 nucleic acid levels (3'LTR) were measured as described. (D) MDDCs were challenged with HIV/Vpx for 6 hr in the absence or presence of NVP (5μM) or Ral (5μM). Fold enhancement of HIV-1 sequences (3'LTR) from the PQBP1 IP over the control normal IgG IP measured by qPCR. (E) Knock down efficiency of PQBP1 in THP-1 cells expressing shRNAs against a scramble control or the PQBP1 protein was measured by qRT-PCR (left) and western blot (right). (F) Infected MDDC lysates were separated into soluble non-chromatin (SNC) and chromatin (C) fractions. DNA isolated from each fraction, a plasmid encoding HIV-1 proviral DNA (circular or linearized) was electroporated into THP-1 cells harboring a shRNA control or shRNA targeting PQBP1 and ISG54 expression was measured. (G) The co-immunoprecipitated DNA as in (A) was eluted and electroporated into PMA-differentiated shControl or shPQBP1 THP-1 cells and ISG54 expression was assessed. (H) MDDCs in the absence or presence of NVP were infected with HIV/Vpx for 6 hr and treated as in (A), and the immunoprecipitated DNA was electroporated into PMA-THP and ISG54 expression was assessed. Data in panels (A) - (D) are shown as the average ± SD of technical quadruplicates. Panels (A) – (D) are representative of at least three independent experiments and (F) – (H) are representative of two independent experiments. *p < 0.05 as determined by 1 way ANOVA with Tukey's post-test. See also Figure S3.
Figure 4. PQBP1 is a specific sensor of retroviral DNA
(A) MDDCs were transfected with a control siRNA or siPQBP1 and challenged with HIV/Vpx, B-DNA, or HT-DNA, then ISG54 expression was measured. (B) PMA-differentiated shControl or shPQBP1 THP-1 cells were infected with HIV/Vpx or transfected with HT-DNA for 8 hr, followed by measuring ISG54 induction. Both infection and transfection were performed in parallel. (C) PMA-differentiated shControl or shPQBP1 THP-1 cells were infected with MHV-68 virus at an MOI 5, and ISG54 induction was measured. (D) THP-1 were challenged with HIV/Vpx or MHV-68 for 6 hr, followed by formaldehyde cross-linking. The abundance of HIV-1- or MHV-68-encoded nucleic acids or mitochondrial DNAs associated with the PQBP1 IP was evaluated (SI Table 2). (E) PMA-differentiated shControl or shPQBP1 THP-1 cells were infected with VSV-G pseudotyped FIV or EIAV, followed by measuring ISG54 induction. (F) CRISPR PQBP1 clones were infected with increasing MOIs of FIV (left) or MHV-68 (right) and ISG54 induction was measured. All data except (D) are shown as the average ± SD of biological triplicates and are representative of at least three independent experiments. For the THP-1 infected with MHV-68 in (D), the lysates were pooled from 6 biological replicates. *p < 0.05 as determined by 1 way ANOVA with Tukey's post-test (A, B), or by an unpaired Student's t-test (E). See also Figure S4.
Figure 5. PQBP1 functions upstream of cGAS and regulates cGAS-STING-dependent IRF3 activation upon retroviral infection
(A) MDDCs were transfected with siRNAs targeting cGAS or a scramble control, challenged with either HIV/Vpx or HT-DNA for 8 hr, and assayed for relative ISG54 mRNA levels. (B) Synthetic cGAMP was incubated with PFO-treated shControl or shPQBP1 THP-1 cells and induction of p-IRF3 was analyzed. (C) Synthetic cGAMP was incubated with PFO-treated shcGAS, shSTING and shLuciferase THP-1 cells and induction of p-IRF3 was analyzed. Knock down efficiency of the target proteins in shcGAS and shSTING lines compared to the shLuc line is shown (right). (D-E) cGAMP levels in the lysates of MDDCs transfected with indicated siRNAs (top) or PMA-differentiated shRNA THP-1 cells (bottom) were assayed prior to and after HIV/Vpx infection. Infected cellular lysates were incubated with PFO-treated THP-1 cells, followed by western blot analysis to detect p-IRF3 (D) or directly quantified using LC-MS (E). The level of cGAMP was graphed as a % increase in HIV-1 infected cells over the untreated samples either in the presence (si/shControl) or absence of PQBP1 protein (si/shPQBP1). (F) cGAMP levels in the lysates of MDDCs were assayed prior to and after HT-DNA stimulation. cGAMP was directly quantified using LC-MS. The level of cGAMP was graphed as a % increase in HT-DNA stimulated cells over the untreated samples either in the presence (siControl) or absence of cGAS protein (sicGAS). Knock down efficiency of cGAS protein in the lysate is shown at the bottom. Panel (A) is shown as the average ± SD of biological triplicates. The data in (A)– ((E)) are representative of at least two independent experiments. In panel (D) - (F) the data were from 6 biological replicates pooled to generate the lysate. For the MDDCs, a single donor was used for the biological replicates in a single experiment. The data are representative of four independent experiments. For the experiments using THP-1, two independent experiments were performed. *p < 0.05 as determined by an unpaired Student's t-test. See also Figure S5.
Figure 6. The WW domain of PQBP1 directs interaction with cGAS
(A) MDDCs (left) and THP-1 (right) were fractionated, as shown in schematic (left), yielding a soluble cytosolic component (S1), a membrane- and organelle-associated component (S2) and a chromatin component (P2). Presence of cGAS, STING, DDX3X, Histone-H3 and PQBP1 in specific fractions were measured by western blot. (B) THP-1 cells stably expressing V5-tagged WT-PQBP1 were lysed and immunoprecipitated for V5 or a normal serum IgG, then probed for endogenous cGAS. (C) HEK293T cells were transfected with eYFP-tagged PQBP1 or eYFP, V5-tagged cGAS and/or FLAG-tagged STING. The cells were lysed and eYFP was immunoprecipitated, followed by a western blot for V5 or FLAG. (D) HEK293T cell lysates expressing V5-tagged cGAS and eYFP-tagged wild type (WT), C-terminal truncation mutant (delC) or WW domain mutant (WW) PQBP1 were immunoprecipitated with an antibody against eYFP and probed for V5. (A) – (D) are representative of at least two independent experiments.
Figure 7. The C-terminal domain of PQBP1 binds HIV-1 DNA and is required for the innate immune response to HIV-1 infection
(A) Recombinant GST or GST-tagged PQBP1 proteins were incubated with either an HIV-1 proviral plasmid or extra-chromosomal nucleic acids (non-chromatin DNA) derived from HIV/Vpx infected MDDCs, and were then immunoprecipitated by GST antibody. The relative abundance of HIV-1 DNA in the input (left) and the IPs (right) is indicated, as measured by qPCR against HIV-1 3’ LTR sequences. (B) Recombinant GST or GST-PQBP1 proteins (WT, WW mutant or C-terminal truncation (1-176) mutant) were incubated with non-chromatin DNA, as in (A), followed by immunoprecipitation with a PQBP1 antibody. The graph at left depicts the relative binding of HIV-1 DNA to the mutant proteins compared to the wild type PQBP1 protein. The blots at right show input and IP'ed GST and/or GST-PQBP1 proteins used in the assay. – indicates GST-PQBP1 proteins, both wild type and mutants. “*” indicates a non-specific band. (C) cPQBP1 THP-1 cells reconstituted with V5-tagged wild type (WT), WW or delC PQBP1 were fractionated as in Figure 6A, and the subcellular fractions were analyzed for the indicated proteins. (D) cPQBP1 THP-1 cells, stably reconstituted with wild type (WT), delC or WW mutant PQBP1, were infected with HIV/Vpx for 6 hr and were subjected to formaldehyde cross-linking followed by assaying for fold enrichment of HIV-1 DNA immunoprecipitated with the reconstituted PQBP1 protein over normal IgG control. Levels of PQBP1 protein immunoprecipitated are shown by western blot (bottom). (E) cPQBP1 THP-1 cells, stably reconstituted with wild type (WT), delC or WW mutant PQBP1, were infected with HIV/Vpx and fold induction of ISG54 over an uninfected sample was measured by qRT-PCR. All panels are representative of at least two independent experiments. (F) MDDCs from two healthy donors and two male Renpenning syndrome patients were challenged with HIV/Vpx. ISG54 induction after 8 hr of infection was evaluated by qRT-PCR and normalized to TBP mRNA levels. ISG54 values for each independent biological replicate are shown. p < 0.0001 (two way ANOVA) comparing HIV-1 challenged samples of healthy and Renpenning cells for each matched set. See also Figure S6.
Similar articles
- Role of PQBP1 in Pathogen Recognition-Impact on Innate Immunity.
Wiench L, Rizzo D, Sinay Z, Nacsa Z, Fuchs NV, König R. Wiench L, et al. Viruses. 2024 Aug 21;16(8):1340. doi: 10.3390/v16081340. Viruses. 2024. PMID: 39205314 Free PMC article. Review. - Recognition of HIV-1 capsid by PQBP1 licenses an innate immune sensing of nascent HIV-1 DNA.
Yoh SM, Mamede JI, Lau D, Ahn N, Sánchez-Aparicio MT, Temple J, Tuckwell A, Fuchs NV, Cianci GC, Riva L, Curry H, Yin X, Gambut S, Simons LM, Hultquist JF, König R, Xiong Y, García-Sastre A, Böcking T, Hope TJ, Chanda SK. Yoh SM, et al. Mol Cell. 2022 Aug 4;82(15):2871-2884.e6. doi: 10.1016/j.molcel.2022.06.010. Epub 2022 Jul 8. Mol Cell. 2022. PMID: 35809572 Free PMC article. - NONO Detects the Nuclear HIV Capsid to Promote cGAS-Mediated Innate Immune Activation.
Lahaye X, Gentili M, Silvin A, Conrad C, Picard L, Jouve M, Zueva E, Maurin M, Nadalin F, Knott GJ, Zhao B, Du F, Rio M, Amiel J, Fox AH, Li P, Etienne L, Bond CS, Colleaux L, Manel N. Lahaye X, et al. Cell. 2018 Oct 4;175(2):488-501.e22. doi: 10.1016/j.cell.2018.08.062. Epub 2018 Sep 27. Cell. 2018. PMID: 30270045 - HIV-1 Activation of Innate Immunity Depends Strongly on the Intracellular Level of TREX1 and Sensing of Incomplete Reverse Transcription Products.
Kumar S, Morrison JH, Dingli D, Poeschla E. Kumar S, et al. J Virol. 2018 Jul 31;92(16):e00001-18. doi: 10.1128/JVI.00001-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29769349 Free PMC article. - First Korean Case of Renpenning Syndrome with Novel Mutation in PQBP1 Diagnosed by Targeted Exome Sequencing, and Literature Review.
Jeong HI, Yang A, Kim J, Jang JH, Cho SY, Jin DK. Jeong HI, et al. Ann Clin Lab Sci. 2018 Jul;48(4):522-527. Ann Clin Lab Sci. 2018. PMID: 30143497 Review.
Cited by
- Intron retention as an excellent marker for diagnosing depression and for discovering new potential pathways for drug intervention.
Okada N, Oshima K, Maruko A, Sekine M, Ito N, Wakasugi A, Mori E, Odaguchi H, Kobayashi Y. Okada N, et al. Front Psychiatry. 2024 Sep 19;15:1450708. doi: 10.3389/fpsyt.2024.1450708. eCollection 2024. Front Psychiatry. 2024. PMID: 39364384 Free PMC article. - Protein-protein interactions in cGAS-STING pathway: a medicinal chemistry perspective.
Zhang SD, Li H, Zhou YL, Liu XC, Li DC, Hao CF, You QD, Xu XL. Zhang SD, et al. Future Med Chem. 2024;16(17):1801-1820. doi: 10.1080/17568919.2024.2383164. Epub 2024 Sep 12. Future Med Chem. 2024. PMID: 39263789 Review. - Role of PQBP1 in Pathogen Recognition-Impact on Innate Immunity.
Wiench L, Rizzo D, Sinay Z, Nacsa Z, Fuchs NV, König R. Wiench L, et al. Viruses. 2024 Aug 21;16(8):1340. doi: 10.3390/v16081340. Viruses. 2024. PMID: 39205314 Free PMC article. Review. - Help or Hinder: Protein Host Factors That Impact HIV-1 Replication.
Moezpoor MR, Stevenson M. Moezpoor MR, et al. Viruses. 2024 Aug 10;16(8):1281. doi: 10.3390/v16081281. Viruses. 2024. PMID: 39205255 Free PMC article. Review. - PQBP3 prevents senescence by suppressing PSME3-mediated proteasomal Lamin B1 degradation.
Yoshioka Y, Huang Y, Jin X, Ngo KX, Kumaki T, Jin M, Toyoda S, Takayama S, Inotsume M, Fujita K, Homma H, Ando T, Tanaka H, Okazawa H. Yoshioka Y, et al. EMBO J. 2024 Sep;43(18):3968-3999. doi: 10.1038/s44318-024-00192-4. Epub 2024 Aug 5. EMBO J. 2024. PMID: 39103492 Free PMC article.
References
- Arad U. Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. BioTechniques. 1998;24:760–762. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- R01 AI105184/AI/NIAID NIH HHS/United States
- P30 CA030199/CA/NCI NIH HHS/United States
- U19 AI106754/AI/NIAID NIH HHS/United States
- P50 GM085764/GM/NIGMS NIH HHS/United States
- U24 DK097153/DK/NIDDK NIH HHS/United States
- R01 DA033773/DA/NIDA NIH HHS/United States
- P01 CA177322/CA/NCI NIH HHS/United States
- P01 AI090935/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials