Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria - PubMed (original) (raw)
Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria
Ido Yosef et al. Proc Natl Acad Sci U S A. 2015.
Abstract
The increasing threat of pathogen resistance to antibiotics requires the development of novel antimicrobial strategies. Here we present a proof of concept for a genetic strategy that aims to sensitize bacteria to antibiotics and selectively kill antibiotic-resistant bacteria. We use temperate phages to deliver a functional clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated (Cas) system into the genome of antibiotic-resistant bacteria. The delivered CRISPR-Cas system destroys both antibiotic resistance-conferring plasmids and genetically modified lytic phages. This linkage between antibiotic sensitization and protection from lytic phages is a key feature of the strategy. It allows programming of lytic phages to kill only antibiotic-resistant bacteria while protecting antibiotic-sensitized bacteria. Phages designed according to this strategy may be used on hospital surfaces and hand sanitizers to facilitate replacement of antibiotic-resistant pathogens with sensitive ones.
Keywords: CRISPR-Cas; ex vivo treatment; lysogenization; positive selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
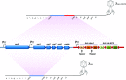
Fig. 1.
Schematics of the lysogenizing phages. The CRISPR-associated genes cas3, cse1, cse2, cas7, cas5, and cas6e (blue) were inserted in place of nucleotides at position 19,014–27,480 of the λ chromosome (National Center for Biotechnology Information Reference Sequence: NC_001416.1) yielding the control lysogenizing phage λcas (Lower). The λcas-CRISPR phage (Upper) encodes, in addition to the cas genes, a CRISPR array with spacers targeting the genes ndm-1 (N1, N2, N3) and ctx-M-15 (C1, C2, C3). PT7, T7 promoter.
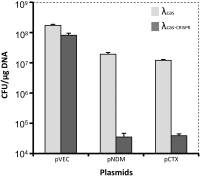
Fig. 2.
Lysogenization effect on transformation of antibiotic resistance plasmids. E. coli K-12 were lysogenized with λcas (light gray bars) or λcas-CRISPR (dark gray bars). These lysogens were transformed with a control (pVEC), ndm-1 (pNDM), or ctx-M-15 (pCTX) encoding plasmids and plated on agar plates supplemented with streptomycin. Bars represent average and SD of the number of CFUs per microgram plasmid counted after plating serial dilutions of the cultures in three independent experiments.
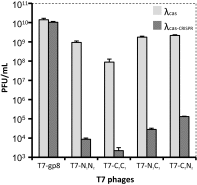
Fig. 3.
Lysogenization effect on protection against lytic phages. E. coli K-12 were lysogenized with λcas (light gray bars) or λcas-CRISPR (dark gray bars). These lysogens were infected with a control T7-gp8 lacking targeted protospacers, or with T7 phages encoding two protospacers from ndm-1 (T7-N1N2) or two protospacers from ctx-M-15 (T7-C2C1) or one spacer from each gene (T7-N1C1 and T7-C2N2). Bars represent average and SD of the number of plaque-forming units (PFUs) per milliliter counted after plating serial dilutions of the phages in three independent experiments.
Fig. 4.
Enrichment of antibiotic-sensitized bacteria by lytic phages. (A) Schematics of the procedure to enrich for antibiotic-sensitized bacteria. A bacterial culture is mixed with lysogenizing phages, resulting in both lysogens and nonlysogens in the culture. Lysogens are both antibiotic sensitized and phage resistant as the CRISPR-Cas system degrades the antibiotic resistance-conferring plasmid and the lytic-phage chromosome. The treated culture is inoculated on agar-containing lytic phages that selectively kill the nonlysogens and thus enrich for antibiotic-sensitized bacteria. (B) Enrichment of phage-resistant E. coli. E. coli K-12 harboring a control (pVEC), ndm-1 (pNDM), ctx-M-15 (pCTX), or ndm-1 + ctx-M-15 (pNDM*/pCTX) encoding plasmids were treated with λcas (light gray bars) or λcas-CRISPR (dark gray bars) and plated on T7-N1C1–coated plates as shown in the scheme presented in A. Bars represent average and SD of the number of surviving CFUs per milliliter counted in three independent experiments. (C) Enrichment of antibiotic-sensitive E. coli. Surviving colonies (20–48 CFUs) from each culture described in B were inoculated on plates having or lacking streptomycin or gentamicin. Bars represent percentage and SD from three independent experiments of streptomycin- or gentamicin-sensitive bacteria scored as CFU unable to grow on plates with streptomycin or gentamicin out of the total number of CFU able to grow on plates lacking these antibiotics.
Similar articles
- Phage-delivered sensitisation with subsequent antibiotic treatment reveals sustained effect against antimicrobial resistant bacteria.
Liu H, Li H, Liang Y, Du X, Yang C, Yang L, Xie J, Zhao R, Tong Y, Qiu S, Song H. Liu H, et al. Theranostics. 2020 May 15;10(14):6310-6321. doi: 10.7150/thno.42573. eCollection 2020. Theranostics. 2020. PMID: 32483454 Free PMC article. - Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting.
Goldberg GW, Jiang W, Bikard D, Marraffini LA. Goldberg GW, et al. Nature. 2014 Oct 30;514(7524):633-7. doi: 10.1038/nature13637. Epub 2014 Aug 31. Nature. 2014. PMID: 25174707 Free PMC article. - Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes.
Edgar R, Friedman N, Molshanski-Mor S, Qimron U. Edgar R, et al. Appl Environ Microbiol. 2012 Feb;78(3):744-51. doi: 10.1128/AEM.05741-11. Epub 2011 Nov 23. Appl Environ Microbiol. 2012. PMID: 22113912 Free PMC article. - Keeping crispr in check: diverse mechanisms of phage-encoded anti-crisprs.
Trasanidou D, Gerós AS, Mohanraju P, Nieuwenweg AC, Nobrega FL, Staals RHJ. Trasanidou D, et al. FEMS Microbiol Lett. 2019 May 1;366(9):fnz098. doi: 10.1093/femsle/fnz098. FEMS Microbiol Lett. 2019. PMID: 31077304 Free PMC article. Review. - Anti-CRISPR: discovery, mechanism and function.
Pawluk A, Davidson AR, Maxwell KL. Pawluk A, et al. Nat Rev Microbiol. 2018 Jan;16(1):12-17. doi: 10.1038/nrmicro.2017.120. Epub 2017 Oct 24. Nat Rev Microbiol. 2018. PMID: 29062071 Review.
Cited by
- Novel Strategy to Combat Antibiotic Resistance: A Sight into the Combination of CRISPR/Cas9 and Nanoparticles.
Wan F, Draz MS, Gu M, Yu W, Ruan Z, Luo Q. Wan F, et al. Pharmaceutics. 2021 Mar 8;13(3):352. doi: 10.3390/pharmaceutics13030352. Pharmaceutics. 2021. PMID: 33800235 Free PMC article. Review. - Bacterial and viral infections and related inflammatory responses in chronic obstructive pulmonary disease.
D'Anna SE, Maniscalco M, Cappello F, Carone M, Motta A, Balbi B, Ricciardolo FLM, Caramori G, Stefano AD. D'Anna SE, et al. Ann Med. 2021 Dec;53(1):135-150. doi: 10.1080/07853890.2020.1831050. Ann Med. 2021. PMID: 32997525 Free PMC article. Review. - Antimicrobial resistance expansion in pathogens: a review of current mitigation strategies and advances towards innovative therapy.
Adefisoye MA, Olaniran AO. Adefisoye MA, et al. JAC Antimicrob Resist. 2023 Dec 11;5(6):dlad127. doi: 10.1093/jacamr/dlad127. eCollection 2023 Dec. JAC Antimicrob Resist. 2023. PMID: 38089461 Free PMC article. Review. - CRISPR-Cas: biology, mechanisms and relevance.
Hille F, Charpentier E. Hille F, et al. Philos Trans R Soc Lond B Biol Sci. 2016 Nov 5;371(1707):20150496. doi: 10.1098/rstb.2015.0496. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27672148 Free PMC article. Review. - The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation.
Langdon A, Crook N, Dantas G. Langdon A, et al. Genome Med. 2016 Apr 13;8(1):39. doi: 10.1186/s13073-016-0294-z. Genome Med. 2016. PMID: 27074706 Free PMC article. Review.
References
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical