Prevalent high-risk HPV infection and vaginal microbiota in Nigerian women - PubMed (original) (raw)
. 2016 Jan;144(1):123-37.
doi: 10.1017/S0950268815000965. Epub 2015 Jun 11.
B Ma 2, A O Famooto 1, S N Adebamowo 1, R A Offiong 3, O Olaniyan 4, P S Dakum 1, C M Wheeler 5, D Fadrosh 2, H Yang 2, P Gajer 2, R M Brotman 2, J Ravel 2, C A Adebamowo 6
Affiliations
- PMID: 26062721
- PMCID: PMC4659743
- DOI: 10.1017/S0950268815000965
Prevalent high-risk HPV infection and vaginal microbiota in Nigerian women
E O Dareng et al. Epidemiol Infect. 2016 Jan.
Abstract
In this study, we evaluated the association between high-risk human papillomavirus (hrHPV) and the vaginal microbiome. Participants were recruited in Nigeria between April and August 2012. Vaginal bacterial composition was characterized by deep sequencing of barcoded 16S rRNA gene fragments (V4) on Illumina MiSeq and HPV was identified using the Roche Linear Array® HPV genotyping test. We used exact logistic regression models to evaluate the association between community state types (CSTs) of vaginal microbiota and hrHPV infection, weighted UniFrac distances to compare the vaginal microbiota of individuals with prevalent hrHPV to those without prevalent hrHPV infection, and the Linear Discriminant Analysis effect size (LEfSe) algorithm to characterize bacteria associated with prevalent hrHPV infection. We observed four CSTs: CST IV-B with a low relative abundance of Lactobacillus spp. in 50% of participants; CST III (dominated by L. iners) in 39·2%; CST I (dominated by L. crispatus) in 7·9%; and CST VI (dominated by proteobacteria) in 2·9% of participants. LEfSe analysis suggested an association between prevalent hrHPV infection and a decreased abundance of Lactobacillus sp. with increased abundance of anaerobes particularly of the genera Prevotella and Leptotrichia in HIV-negative women (P < 0·05). These results are hypothesis generating and further studies are required.
Keywords: HIV/AIDS; human papilloma virus (HPV); public health.
Conflict of interest statement
None.
Figures
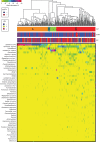
Fig. 1.
Heat map of relative abundance for the 50 most abundant bacterial taxa found in the vaginal bacterial communities of all participants in the study. Ward linkage clustering was used to cluster samples based on their Jensen–Shannon distance calculated in the
vegan
package in R [44]. Identified community state types (CSTs) are labelled as I, III, and IV, according to the previous naming convention [51]. hrHPV, High-risk human papillomavirus.
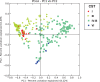
Fig. 2.
Weighted UniFrac principal coordinates analysis (PCoA) plot comparing sample distribution belonging to different community state types (CSTs). See Figure 1 for sample CST assignments used in this figure.
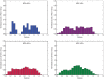
Fig. 3.
Histogram of weighted UniFrac distance between samples by human papillomavirus (HPV)/HIV metadata. The distribution of distance between samples of HPV + /HIV–, HPV–/HIV–, HPV + /HIV + , and HPV–/HIV+ women is shown.
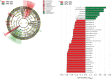
Fig. 4.
(a) Cladogram representing the taxonomic hierarchical structure of the identified phylotype biomarkers, generated using LEfSe [47]. Phylotype biomarkers are identified comparing samples collected from HIV–/HPV– and HIV–/HPV+ participants. Each filled circle represents one biomarker. Red, phylotypes statistically overrepresented under the condition of HPV + /HIV–; green, phylotypes overrepresented under the condition of HPV–/HIV–; yellow, phylotypes for which relative abundance is not significantly different between the two conditions. The diameter of each circle is proportional to the phylotype's effect size, phylum and class are indicated in their names on the cladogram and the order, family, or genera are given in the key. (b) Identified phylotype biomarkers ranked by effect size in HIV– women. The phylotype biomarkers are identified as being significantly abundant comparing samples collected from HPV– and HPV+ women with an alpha value <0·05. The graph was generated using the LEfSe program. The phylotypes are ranked according to their effect size that are associated with different conditions with the highest median. The Linear Discriminant Analysis (LDA) score [47] at the log10 scale is indicated at the bottom. The greater the LDA score is, the more significant the phylotype biomarker is in the comparison.
Similar articles
- Human papillomavirus molecular prevalence in south China and the impact on vaginal microbiome of unvaccinated women.
Wang T, Li W, Cai M, Ji S, Wang Y, Huang N, Jiang Y, Zhang Z. Wang T, et al. mSystems. 2024 Sep 17;9(9):e0073824. doi: 10.1128/msystems.00738-24. Epub 2024 Aug 9. mSystems. 2024. PMID: 39120153 Free PMC article. - Association between the vaginal microbiome and high-risk human papillomavirus infection in pregnant Chinese women.
Chen Y, Hong Z, Wang W, Gu L, Gao H, Qiu L, Di W. Chen Y, et al. BMC Infect Dis. 2019 Aug 1;19(1):677. doi: 10.1186/s12879-019-4279-6. BMC Infect Dis. 2019. PMID: 31370796 Free PMC article. - Human papillomavirus infection and cervical intraepithelial neoplasia progression are associated with increased vaginal microbiome diversity in a Chinese cohort.
Chen Y, Qiu X, Wang W, Li D, Wu A, Hong Z, Di W, Qiu L. Chen Y, et al. BMC Infect Dis. 2020 Aug 26;20(1):629. doi: 10.1186/s12879-020-05324-9. BMC Infect Dis. 2020. PMID: 32842982 Free PMC article. - The vaginal microbiota, human papillomavirus and cervical dysplasia: a systematic review and network meta-analysis.
Norenhag J, Du J, Olovsson M, Verstraelen H, Engstrand L, Brusselaers N. Norenhag J, et al. BJOG. 2020 Jan;127(2):171-180. doi: 10.1111/1471-0528.15854. Epub 2019 Jul 17. BJOG. 2020. PMID: 31237400
Cited by
- The vaginal metabolome and microbiota of cervical HPV-positive and HPV-negative women: a cross-sectional analysis.
Borgogna JC, Shardell MD, Santori EK, Nelson TM, Rath JM, Glover ED, Ravel J, Gravitt PE, Yeoman CJ, Brotman RM. Borgogna JC, et al. BJOG. 2020 Jan;127(2):182-192. doi: 10.1111/1471-0528.15981. Epub 2019 Nov 20. BJOG. 2020. PMID: 31749298 Free PMC article. - Molecular characterisation of genital human papillomavirus among women in Southwestern, Nigeria.
Nejo YT, Olaleye DO, Odaibo GN. Nejo YT, et al. PLoS One. 2019 Nov 4;14(11):e0224748. doi: 10.1371/journal.pone.0224748. eCollection 2019. PLoS One. 2019. PMID: 31682636 Free PMC article. - Vaginal dysbiosis seems associated with hrHPV infection in women attending the Dutch Cervical Cancer Screening Program.
Loonen AJM, Verhagen F, Luijten-de Vrije I, Lentjes-Beer M, Huijsmans CJ, van den Brule AJC. Loonen AJM, et al. Front Cell Infect Microbiol. 2024 Mar 13;14:1330844. doi: 10.3389/fcimb.2024.1330844. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38544527 Free PMC article. - Vaginal microbiota stability over 18 months in young student women in France.
Tamarelle J, Thiébaut ACM, de Barbeyrac B, Bébéar C, Bourret A, Fauconnier A, Ravel J, Delarocque-Astagneau E; i-Predict study group. Tamarelle J, et al. Eur J Clin Microbiol Infect Dis. 2024 Dec;43(12):2277-2292. doi: 10.1007/s10096-024-04943-3. Epub 2024 Sep 20. Eur J Clin Microbiol Infect Dis. 2024. PMID: 39302529 - Public human microbiome data are dominated by highly developed countries.
Abdill RJ, Adamowicz EM, Blekhman R. Abdill RJ, et al. PLoS Biol. 2022 Feb 15;20(2):e3001536. doi: 10.1371/journal.pbio.3001536. eCollection 2022 Feb. PLoS Biol. 2022. PMID: 35167588 Free PMC article.
References
- Ferlay J, et al. GLOBOCAN 2012 v. 1.0, Cancer incidence and mortality worldwide: IARC Cancer Base No. 11. Lyon, France: International Agency for Research on Cancer, 2013.
- Parkin DM, et al. Cancer in Africa 2012. Cancer Epidemiology, Biomarkers and Prevention 2014; 23: 953–966. - PubMed
- Jemal A, et al. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiology, Biomarkers and Prevention 2010; 19: 1893–1907. - PubMed
- Galloway D, et al. IARC monographs on the evaluation of carcinogenic risks to humans: human papilloma viruses, volume 90. World Health Organization, International Agency for Research on Cancer, 2007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1U54HG006947-01A1/HG/NHGRI NIH HHS/United States
- U54 HG006947/HG/NHGRI NIH HHS/United States
- U19 AI084044/AI/NIAID NIH HHS/United States
- 1D43CA153792/CA/NCI NIH HHS/United States
- U19 AI084081/AI/NIAID NIH HHS/United States
- D43 CA153792/CA/NCI NIH HHS/United States
- U19AI084044/AI/NIAID NIH HHS/United States
- K01AI080974/AI/NIAID NIH HHS/United States
- U19 AI113187/AI/NIAID NIH HHS/United States
- K01 AI080974/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous