Two Mutations Were Critical for Bat-to-Human Transmission of Middle East Respiratory Syndrome Coronavirus - PubMed (original) (raw)
Two Mutations Were Critical for Bat-to-Human Transmission of Middle East Respiratory Syndrome Coronavirus
Yang Yang et al. J Virol. 2015 Sep.
Abstract
To understand how Middle East respiratory syndrome coronavirus (MERS-CoV) transmitted from bats to humans, we compared the virus surface spikes of MERS-CoV and a related bat coronavirus, HKU4. Although HKU4 spike cannot mediate viral entry into human cells, two mutations enabled it to do so by allowing it to be activated by human proteases. These mutations are present in MERS-CoV spike, explaining why MERS-CoV infects human cells. These mutations therefore played critical roles in the bat-to-human transmission of MERS-CoV, either directly or through intermediate hosts.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
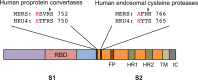
FIG 1
Domain structure of MERS-CoV and HKU4 spike proteins. The spikes contain a receptor-binding S1 subunit, a membrane-fusion S2 subunit, a transmembrane anchor (TM), and an intracellular tail (IC). S1 contains the receptor-binding domain (RBD) that binds DPP4 receptor. S2 contains the fusion peptide (FP), heptad repeat 1 (HR1), and heptad repeat 2 (HR2), all of which are essential structural elements for the membrane fusion process. The S1/S2 boundary in MERS-CoV spike (defined as the region between the RBD and the fusion peptide) contains one established human protease motif that is recognized by proprotein convertases (hPPC) (15, 16); it also contains one established human protease motif that is recognized by endosomal cysteine proteases (hECP) (17, 18). Sequence alignments of these regions in MERS-CoV and HKU4 spikes (GenBank accession no. AFS88936.1 for MERS-CoV spike; ABN10839.1 for HKU4 spike) are shown, with the critical residue differences labeled in red. Arrows indicate the predicted sites of cleavage by human proteases in MERS-CoV spike.
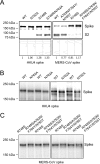
FIG 2
Characterization of two protease motifs in MERS-CoV and HKU4 spike proteins. (A) Western blot analysis of HKU4 and MERS-CoV spikes in pseudovirus particles. Retroviruses pseudotyped with HKU4 spike (i.e., HKU4 pseudoviruses) or MERS-CoV spike (i.e., MERS-CoV pseudoviruses) were prepared in HEK293T (human embryonic kidney) cells as previously described (14). The incorporations of wild-type (WT) and mutant HKU4 and MERS-CoV spikes into pseudovirus particles were measured by Western blotting using antibody against their C-terminal C9 tags. Plots below the Western blot images correspond to quantifications of the band intensities from the cleaved and uncleaved spikes combined. The numbers below the plots indicate the relative amounts of spikes incorporated into pseudovirus particles compared to wild-type HKU4 and MERS-CoV spikes, respectively. All quantifications were done using ImageJ software (National Institutes of Health). (B) Glycosylation state of HKU4 spike at the hECP motif. HEK293T cells exogenously expressing wild-type (WT) and mutant HKU4 spikes were lysed and subjected to Western blot analysis. To improve the separation of high-molecular-mass spikes, 3 to 8% NuPAGE Tris-acetate gels (Life Technologies) were used for gel electrophoresis. To improve the accuracy of the result, each of the mutant and wild-type spikes was run in two lanes that alternated between samples. The experiment was also repeated in a separate gel. Compared with wild-type HKU4 spike, the downward shift in the band of HKU4 spike bearing the mutated hECP motif (i.e., mutation N762A) is consistent with the removal of glycosylation. (C) Glycosylation state of MERS-CoV spike at the hECP motif. Compared with MERS-CoV spike bearing the reengineered hPPC motif (i.e., mutation R748S), the upward shift in the band of MERS-CoV spike bearing both the reengineered hPPC and hECP motifs (i.e., mutations R748S/A763N/F764Y/N765T) indicates the introduction of glycosylation.
FIG 3
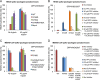
HKU4- and MERS-CoV-spike-mediated pseudovirus entry in human cells. (A) HKU4 pseudoviruses bearing no mutation, the reengineered hPPC motif (S746R), the reengineered hECP motif (N762A), or both of the reengineered motifs (S746R/N762A) were prepared in HEK293T (human embryonic kidney) cells and were then used to infect HEK293T cells exogenously expressing human DPP4 (GenBank accession no. NP_001926.2). The infections were carried out in the presence or absence of exogenous trypsin. The pseudovirus entry efficiencies were characterized by analysis of the levels of luciferase activity accompanying the entry and were normalized against the relative amounts of spikes incorporated into pseudovirus particles (Fig. 2A). The pseudovirus entry mediated by HKU4 spike bearing both of the reengineered motifs in the absence of exogenous trypsin was taken as 100%. (B) Pseudovirus-producing HEK293T cells were treated with proprotein convertase (PPC) inhibitor (dec-RVKR-CMK) 5 h after transfection of plasmids that encode HKU4 spike containing the reengineered hPPC motif (S746R). Pseudovirus-targeting HEK293T cells were treated with endosomal cysteine protease (ECP) inhibitor (E-64d) before being infected by HKU4 pseudoviruses bearing the reengineered hECP motif (N762A). Both of the inhibitors were used for HKU4 pseudoviruses bearing both of the mutations. (C) MERS-CoV pseudoviruses bearing no mutation, the mutated hPPC motif (R748S), the mutated hECP motif (A763N/F764Y/N765T), or both of the mutated motifs (R748S/A763N/F764Y/N765T) were used to infect HEK293T cells exogenously expressing human DPP4. The pseudovirus entry mediated by the wild-type MERS-CoV spike in the absence of exogenous trypsin was taken as 100%. (D) Pseudovirus-producing HEK293T cells were treated with dec-RVKR-CMK 5 h after transfection of plasmids that encode MERS-CoV spike containing the mutated hPPC motif (R748S). Pseudovirus-targeting HEK293T cells were treated with E-64d before being infected by MERS-CoV pseudoviruses bearing the mutated hECP motif (A763N/F764Y/N765T). Both of the inhibitors were used for MERS-CoV pseudoviruses bearing both of the mutated motifs. Error bars indicate standard errors of the means (SEM) (n = 4).
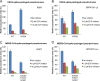
FIG 4
HKU4- and MERS-CoV-spike-mediated pseudovirus entry into bat cells. (A) HKU4 pseudoviruses bearing no mutation or the reengineered hECP motif (N762A) were prepared in HEK293T cells and were then used to infect RSKT cells endogenously expressing bat DPP4 (Rhinolophus sinicus bat kidney cells [24]). The cells were pretreated with indicated concentrations of ECP inhibitor (E-64d) before being infected by pseudoviruses. (B) The same HKU4 pseudoviruses were used to infect Tb1-Lu cells (Tadarida brasiliensis bat lung cells) exogenously expressing bat DPP4 (GenBank accession no. KC249974). (C) MERS-CoV pseudoviruses bearing no mutation or the mutant hECP motif (A763N/F764Y/N765T) were prepared in HEK293T cells and were then used to infect RSKT cells. (D) The same MERS-CoV pseudoviruses were used to infect Tb1-Lu cells. The pseudovirus entry efficiencies were characterized by analysis of the levels of luciferase activity accompanying the entry and were normalized against the relative amounts of spike proteins incorporated into pseudovirus particles. The pseudovirus entry mediated by the wild-type HKU4 spike (for panels A and B) or wild-type MERS-CoV spike (for panels C and D) in the absence of the inhibitor was taken as 100%. Error bars indicate SEM (n = 4).
Similar articles
- Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26.
Wang Q, Qi J, Yuan Y, Xuan Y, Han P, Wan Y, Ji W, Li Y, Wu Y, Wang J, Iwamoto A, Woo PC, Yuen KY, Yan J, Lu G, Gao GF. Wang Q, et al. Cell Host Microbe. 2014 Sep 10;16(3):328-37. doi: 10.1016/j.chom.2014.08.009. Cell Host Microbe. 2014. PMID: 25211075 Free PMC article. - Permissivity of Dipeptidyl Peptidase 4 Orthologs to Middle East Respiratory Syndrome Coronavirus Is Governed by Glycosylation and Other Complex Determinants.
Peck KM, Scobey T, Swanstrom J, Jensen KL, Burch CL, Baric RS, Heise MT. Peck KM, et al. J Virol. 2017 Sep 12;91(19):e00534-17. doi: 10.1128/JVI.00534-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28747502 Free PMC article. - Rapid detection of MERS coronavirus-like viruses in bats: pote1ntial for tracking MERS coronavirus transmission and animal origin.
Woo PCY, Lau SKP, Chen Y, Wong EYM, Chan KH, Chen H, Zhang L, Xia N, Yuen KY. Woo PCY, et al. Emerg Microbes Infect. 2018 Mar 7;7(1):18. doi: 10.1038/s41426-017-0016-7. Emerg Microbes Infect. 2018. PMID: 29511173 Free PMC article. - Coronavirus Host Range Expansion and Middle East Respiratory Syndrome Coronavirus Emergence: Biochemical Mechanisms and Evolutionary Perspectives.
Peck KM, Burch CL, Heise MT, Baric RS. Peck KM, et al. Annu Rev Virol. 2015 Nov;2(1):95-117. doi: 10.1146/annurev-virology-100114-055029. Epub 2015 Aug 7. Annu Rev Virol. 2015. PMID: 26958908 Review. - [A novel coronavirus, MERS-CoV].
Mizutani T. Mizutani T. Uirusu. 2013;63(1):1-6. doi: 10.2222/jsv.63.1. Uirusu. 2013. PMID: 24769571 Review. Japanese.
Cited by
- Unraveling the stability landscape of mutations in the SARS-CoV-2 receptor-binding domain.
Smaoui MR, Yahyaoui H. Smaoui MR, et al. Sci Rep. 2021 Apr 28;11(1):9166. doi: 10.1038/s41598-021-88696-5. Sci Rep. 2021. PMID: 33911163 Free PMC article. - MERS-CoV spike protein: Targets for vaccines and therapeutics.
Wang Q, Wong G, Lu G, Yan J, Gao GF. Wang Q, et al. Antiviral Res. 2016 Sep;133:165-77. doi: 10.1016/j.antiviral.2016.07.015. Epub 2016 Jul 26. Antiviral Res. 2016. PMID: 27468951 Free PMC article. Review. - Structural insights into coronavirus entry.
Tortorici MA, Veesler D. Tortorici MA, et al. Adv Virus Res. 2019;105:93-116. doi: 10.1016/bs.aivir.2019.08.002. Epub 2019 Aug 22. Adv Virus Res. 2019. PMID: 31522710 Free PMC article. - In silico comparative genomics of SARS-CoV-2 to determine the source and diversity of the pathogen in Bangladesh.
Shishir TA, Naser IB, Faruque SM. Shishir TA, et al. PLoS One. 2021 Jan 20;16(1):e0245584. doi: 10.1371/journal.pone.0245584. eCollection 2021. PLoS One. 2021. PMID: 33471859 Free PMC article. - Evolutionary Dynamics of MERS-CoV: Potential Recombination, Positive Selection and Transmission.
Zhang Z, Shen L, Gu X. Zhang Z, et al. Sci Rep. 2016 May 4;6:25049. doi: 10.1038/srep25049. Sci Rep. 2016. PMID: 27142087 Free PMC article.
References
- de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish ZA, Perlman S, Poon LL, Snijder EJ, Stephens GM, Woo PC, Zaki AM, Zambon M, Ziebuhr J. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol 87:7790–7792. doi:10.1128/JVI.01244-13. - DOI - PMC - PubMed
- Annan A, Baldwin HJ, Corman VM, Klose SM, Owusu M, Nkrumah EE, Badu EK, Anti P, Agbenyega O, Meyer B, Oppong S, Sarkodie YA, Kalko EK, Lina PH, Godlevska EV, Reusken C, Seebens A, Gloza-Rausch F, Vallo P, Tschapka M, Drosten C, Drexler JF. 2013. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis 19:456–459. doi:10.3201/eid1903.121503. - DOI - PMC - PubMed
- Lau SK, Li KS, Tsang AK, Lam CS, Ahmed S, Chen H, Chan KH, Woo PC, Yuen KY. 2013. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J Virol 87:8638–8650. doi:10.1128/JVI.01055-13. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI089728/AI/NIAID NIH HHS/United States
- R01 AI110700/AI/NIAID NIH HHS/United States
- R01AI089728/AI/NIAID NIH HHS/United States
- R01AI110700/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources