Autophagy and Lipid Droplets in the Liver - PubMed (original) (raw)
Review
Autophagy and Lipid Droplets in the Liver
Nuria Martinez-Lopez et al. Annu Rev Nutr. 2015.
Abstract
Autophagy is a conserved quality-control pathway that degrades cytoplasmic contents in lysosomes. Autophagy degrades lipid droplets through a process termed lipophagy. Starvation and an acute lipid stimulus increase autophagic sequestration of lipid droplets and their degradation in lysosomes. Accordingly, liver-specific deletion of the autophagy gene Atg7 increases hepatic fat content, mimicking the human condition termed nonalcoholic fatty liver disease. In this review, we provide insights into the molecular regulation of lipophagy, discuss fundamental questions related to the mechanisms by which autophagosomes recognize lipid droplets and how ATG proteins regulate membrane curvature for lipid droplet sequestration, and comment on the possibility of cross talk between lipophagy and cytosolic lipases in lipid mobilization. Finally, we discuss the contribution of lipophagy to the pathophysiology of human fatty liver disease. Understanding how lipophagy clears hepatocellular lipid droplets could provide new ways to prevent fatty liver disease, a major epidemic in developed nations.
Keywords: lipases; lipophagy; lysosome; metabolism; steatosis.
Figures
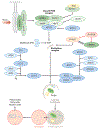
Figure 1
The autophagy pathway. Stress or starvation activates the class III phosphatidylinositol 3-kinases (PI3K) complex that generates phosphatidylinositol 3-phosphates (PI3Ps). PI3Ps recruit autophagy genes (ATGs) to the nucleation complex including UNC51-like kinase 1 (ULK1), an upstream regulator of autophagy. Two conjugation cascades, i.e., the ATG12-ATG5 and light chain 3 (LC3) conjugation cascades, are activated, which results in lipidation of cytosolic LC3-I into membrane-bound LC3-II. LC3-II-positive autophagosomes sequester cytoplasmic cargo that is delivered to the lysosomes, wherein a battery of acid hydrolases degrade engulfed cargo. Products of degradation—amino acids, fatty acids, and nucleic acids—are utilized by the starved cell. Abbreviations: AMBRA1, activating molecule in Beclin1-regulated autophagy; Beclin1, coiled-coil, myosin-like BCL2-interacting protein; Dynein HC, dynein heavy chain; P-mTOR, phospho-mammalian target of rapamycin; P-ULK1, phospho-UNC51-like kinase 1.
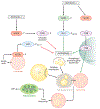
Figure 2
The regulation of lipophagy. The availability of nutrients phosphorylates and activates the mechanistic target of rapamycin (mTOR). It is likely that lysosomal efflux of amino acids also activates mTOR. Active mTOR phosphorylates transcription factor EB (TFEB) and retains it in the cytoplasm, and it decreases autophagy and lysosomal gene expression. Active mTOR also suppresses UNC51-like kinase 1 (ULK1) activity by phosphorylating ULK1 at SER 757. During nutrient depletion, mTOR is inactivated while adenosine monophosphate-activated protein kinase (AMPK) is activated. These events allow ULK1 phosphorylation at SER 555, which results in ULK1 activation. Suppression of mTOR activity also allows TFEB to localize to the nucleus to drive autophagy and lysosomal gene expression. Activation of autophagy begins to sequester and degrade lipid droplets (LDs) in lysosomes. Lysosome-derived free fatty acids are oxidized in the mitochondria for production of adenosine triphosphate (ATP).
Similar articles
- PNPLA3 variant M148 causes resistance to starvation-mediated lipid droplet autophagy in human hepatocytes.
Negoita F, Blomdahl J, Wasserstrom S, Winberg ME, Osmark P, Larsson S, Stenkula KG, Ekstedt M, Kechagias S, Holm C, Jones HA. Negoita F, et al. J Cell Biochem. 2019 Jan;120(1):343-356. doi: 10.1002/jcb.27378. Epub 2018 Sep 1. J Cell Biochem. 2019. PMID: 30171718 - Telemetric control of peripheral lipophagy by hypothalamic autophagy.
Martinez-Lopez N, Singh R. Martinez-Lopez N, et al. Autophagy. 2016 Aug 2;12(8):1404-5. doi: 10.1080/15548627.2016.1185578. Epub 2016 Jun 24. Autophagy. 2016. PMID: 27341145 Free PMC article. - Lipophagy and liver disease: New perspectives to better understanding and therapy.
Zhang Z, Yao Z, Chen Y, Qian L, Jiang S, Zhou J, Shao J, Chen A, Zhang F, Zheng S. Zhang Z, et al. Biomed Pharmacother. 2018 Jan;97:339-348. doi: 10.1016/j.biopha.2017.07.168. Epub 2017 Nov 6. Biomed Pharmacother. 2018. PMID: 29091883 Review. - Involvement of Lipophagy and Chaperone-Mediated Autophagy in the Pathogenesis of Non-Alcoholic Fatty Liver Disease by Regulation of Lipid Droplets.
Mastoridou EM, Goussia AC, Kanavaros P, Charchanti AV. Mastoridou EM, et al. Int J Mol Sci. 2023 Nov 2;24(21):15891. doi: 10.3390/ijms242115891. Int J Mol Sci. 2023. PMID: 37958873 Free PMC article. Review. - ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism.
Sathyanarayan A, Mashek MT, Mashek DG. Sathyanarayan A, et al. Cell Rep. 2017 Apr 4;19(1):1-9. doi: 10.1016/j.celrep.2017.03.026. Cell Rep. 2017. PMID: 28380348 Free PMC article.
Cited by
- Comparative Histological Study of Therapeutic Effect of Mesenchymal Stem Cells versus Mesenchymal Stem Cells Co-Cultured with Liver Tissue on Carbon Tetrachloride-Induced Hepatotoxicity in Adult Male Albino Rats.
Salem BA, ElKaliny HH, El-Hafez AAAA, Sarhan NI. Salem BA, et al. J Microsc Ultrastruct. 2022 Aug 18;11(4):225-236. doi: 10.4103/jmau.jmau_62_21. eCollection 2023 Oct-Dec. J Microsc Ultrastruct. 2022. PMID: 38213650 Free PMC article. - Cholesterol-induced leucine aminopeptidase 3 (LAP3) upregulation inhibits cell autophagy in pathogenesis of NAFLD.
Feng L, Chen Y, Xu K, Li Y, Riaz F, Lu K, Chen Q, Du X, Wu L, Cao D, Li C, Lu S, Li D. Feng L, et al. Aging (Albany NY). 2022 Apr 11;14(7):3259-3275. doi: 10.18632/aging.204011. Epub 2022 Apr 11. Aging (Albany NY). 2022. PMID: 35404840 Free PMC article. - Verapamil induces autophagy to improve liver regeneration in non-alcoholic fatty liver mice.
Lai JL, Lian YE, Wu JY, Wang YD, Bai YN. Lai JL, et al. Adipocyte. 2021 Dec;10(1):532-545. doi: 10.1080/21623945.2021.1983241. Adipocyte. 2021. PMID: 34699301 Free PMC article. - Mitophagy in the Pathogenesis of Liver Diseases.
Ke PY. Ke PY. Cells. 2020 Mar 30;9(4):831. doi: 10.3390/cells9040831. Cells. 2020. PMID: 32235615 Free PMC article. Review. - Regulation of lipophagy in NAFLD by cellular metabolism and CD36.
Samovski D, Abumrad NA. Samovski D, et al. J Lipid Res. 2019 Apr;60(4):755-757. doi: 10.1194/jlr.C093674. Epub 2019 Feb 28. J Lipid Res. 2019. PMID: 30819696 Free PMC article. No abstract available.
References
- Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, et al. 2007. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet 16:2600–15 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K01 DK087776/DK/NIDDK NIH HHS/United States
- DK087776/DK/NIDDK NIH HHS/United States
- AG031782/AG/NIA NIH HHS/United States
- R01 AG043517/AG/NIA NIH HHS/United States
- DK020541/DK/NIDDK NIH HHS/United States
- P60 DK020541/DK/NIDDK NIH HHS/United States
- AG043517/AG/NIA NIH HHS/United States
- P30 DK020541/DK/NIDDK NIH HHS/United States
- RF1 AG043517/AG/NIA NIH HHS/United States
- P01 AG031782/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources