Mutations in DYNC2LI1 disrupt cilia function and cause short rib polydactyly syndrome - PubMed (original) (raw)
Mutations in DYNC2LI1 disrupt cilia function and cause short rib polydactyly syndrome
S Paige Taylor et al. Nat Commun. 2015.
Abstract
The short rib polydactyly syndromes (SRPSs) are a heterogeneous group of autosomal recessive, perinatal lethal skeletal disorders characterized primarily by short, horizontal ribs, short limbs and polydactyly. Mutations in several genes affecting intraflagellar transport (IFT) cause SRPS but they do not account for all cases. Here we identify an additional SRPS gene and further unravel the functional basis for IFT. We perform whole-exome sequencing and identify mutations in a new disease-producing gene, cytoplasmic dynein-2 light intermediate chain 1, DYNC2LI1, segregating with disease in three families. Using primary fibroblasts, we show that DYNC2LI1 is essential for dynein-2 complex stability and that mutations in DYNC2LI1 result in variable length, including hyperelongated, cilia, Hedgehog pathway impairment and ciliary IFT accumulations. The findings in this study expand our understanding of SRPS locus heterogeneity and demonstrate the importance of DYNC2LI1 in dynein-2 complex stability, cilium function, Hedgehog regulation and skeletogenesis.
Conflict of interest statement
Competing Financial Interests
The authors have no competing interests.
Figures
Figure 1
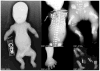
Mutations in DYNC2LI1 cause short rib polydactyly syndrome. Radiographs of the affected probands, R03-303A, R01-013A and R07-628A (International Skeletal Dysplasia Registry reference numbers) showing short long bones, horizontal ribs and long narrow chest, poor mineralization of some skeletal elements (single arrow), metaphyseal irregularity (double arrows), and polydactyly in all the limbs of the affected individuals.
Figure 2
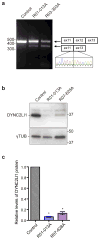
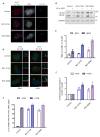
Effect of DYNC2LI1 mutations on splicing and protein stability. (a) RT-PCR of cDNA derived from control and SRPS fibroblasts shows that DYNC2LI1 splice-donor mutations cause in-frame skipping of exon 12. (b) Immunoblotting reveals significant reduction in the amounts of DYNC2LI1 from lysates of affected cells compared with control. Size markers (right, kDa), β-tubulin, loading control. (c) Quantification of average DYNC2LI1 amounts from (b) for each genotype. Values were normalized to control and error bars represent ± s.e.m (3 independent experiments). Statistical analyses performed using the Mann-Whitney test, p<0.05 labeled with *.
Figure 3
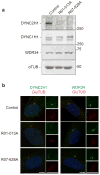
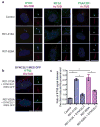
Mutations in DYNC2LI1 decrease the stability of DYNC2H1. (a) Immunoblotting for components of the dynein-2 complex (heavy chain, DYNC2H1; intermediate chain, WDR34) and the heavy chain of the dynein-1 complex (DYNC1H1) reveals a specific reduction of DYNC2H1. (b) Immunofluorescence micrographs of control and SRPS cells stained for GluTUB (red) and WDR34 (green) or DYNC2H1 (green). Cilia (2x) are shown on the right. Scale bar = 5 μm.
Figure 4
Components of the dynein-2 complex are co-expressed in a human cartilage growth plate. (a) Sections from a 14-week human humerus were stained for DYNC2LI1, DYNC2H1 and WDR34. All three proteins were expressed in the perichondrium/periosteum and primary spongiosa (closed arrows, perichondrium/periosteum; open arrows, primary spongiosa). Scale bar = 50 μm.
Figure 5
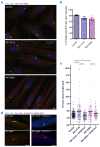
Primary cilia are variable in length and longer in _DYNC2LI1_-mutant cells. (a) Immunofluorescence micrographs of serum-starved control and SRPS fibroblasts stained for ARL13B (green), GluTUB (red) and Hoescht (blue) show variable length and hyperelongated cilia in SRPS cells. (b) Percentage of ciliated cells in (a) (n=150 X 3 independent experiments). (c) Primary cilia length is highly variable in cells with DYNC2LI1 mutations (n>20 X 3 independent experiments). (d) Expression of untagged wild type DYNC2LI1 through an IRES-GFP vector (artificially colored blue) rescues cilia length variability in SRPS cells. Error bars represent ± s.e.m. Statistical analyses performed using Mann-Whitney test, p<0.05 values labeled with *. Scale bar = 5 μm.
Figure 6
DYNC2LI1 mutations delay retrograde IFT and lead to the ciliary accumulation of IFT components. (a) Immunofluorescence micrographs of control and SRPS cells stained as labeled. Cilia (2x) are shown on the right. (b) Expression of untagged wild type DYNC2LI1 through an IRES-GFP vector (artificially colored blue) rescues ciliary accumulation of IFT components. (c) Quantification of IFT88 signal: graph shows the mean ratio of IFT88 signal between the cilium body and its proximal end ± s.e.m (n=20 X 3 independent experiments). Statistical analyses performed using Mann-Whitney test, p<0.05 values labeled with *. Scale bar = 5 μm.
Figure 7
Disruption of the dynein-2 complex leads to aberrant Hedgehog signaling. (a) Immunofluorescence micrographs of control and SRPS cells treated with SAG and stained for GLI3 (red), AcTUB (green), and DAPI (blue). (b) DYNC2LI1/dynein-2 complex activity is required for the exclusion of SMO from the primary cilium in the absence of Hedgehog stimulation. Immunofluorescence micrographs of SAG-stimulated control and SRPS cells stained for SMO (green) and GluTUB (red). (c) Graph shows the mean percentage of ciliated cells with ciliary SMO ± s.e.m. (>100 cells counted X 3 independent experiments). (d) Immunoblotting for GLI3 (full length “GLI3FL”; repressor “GLI3R”) in cell extracts shows increased GLI3FL to GLI3R in SRPS cells. (e) Graph shows the ratio of GLI3FL to GLI3R. (f) Graph shows quantification of total GLI3 amounts (GLI3FL + GLI3R) which is increased in _DYNC2LI1_-mutant fibroblasts. Statistical analyses performed using Mann-Whitney test, p<0.05 is labeled with *. Scale bars = 5 μm.
Similar articles
- DYNC2LI1 mutations broaden the clinical spectrum of dynein-2 defects.
Kessler K, Wunderlich I, Uebe S, Falk NS, Gießl A, Brandstätter JH, Popp B, Klinger P, Ekici AB, Sticht H, Dörr HG, Reis A, Roepman R, Seemanová E, Thiel CT. Kessler K, et al. Sci Rep. 2015 Jul 1;5:11649. doi: 10.1038/srep11649. Sci Rep. 2015. PMID: 26130459 Free PMC article. - Mutations in DYNC2H1, the cytoplasmic dynein 2, heavy chain 1 motor protein gene, cause short-rib polydactyly type I, Saldino-Noonan type.
Badiner N, Taylor SP, Forlenza K, Lachman RS; University of Washington Center for Mendelian Genomics; Bamshad M, Nickerson D, Cohn DH, Krakow D. Badiner N, et al. Clin Genet. 2017 Aug;92(2):158-165. doi: 10.1111/cge.12947. Epub 2017 Mar 13. Clin Genet. 2017. PMID: 27925158 Free PMC article. - An inactivating mutation in intestinal cell kinase, ICK, impairs hedgehog signalling and causes short rib-polydactyly syndrome.
Paige Taylor S, Kunova Bosakova M, Varecha M, Balek L, Barta T, Trantirek L, Jelinkova I, Duran I, Vesela I, Forlenza KN, Martin JH, Hampl A; University of Washington Center for Mendelian Genomics; Bamshad M, Nickerson D, Jaworski ML, Song J, Ko HW, Cohn DH, Krakow D, Krejci P. Paige Taylor S, et al. Hum Mol Genet. 2016 Sep 15;25(18):3998-4011. doi: 10.1093/hmg/ddw240. Epub 2016 Jul 27. Hum Mol Genet. 2016. PMID: 27466187 Free PMC article. - Ciliary Dyneins and Dynein Related Ciliopathies.
Antony D, Brunner HG, Schmidts M. Antony D, et al. Cells. 2021 Jul 25;10(8):1885. doi: 10.3390/cells10081885. Cells. 2021. PMID: 34440654 Free PMC article. Review. - Ciliary disorder of the skeleton.
Huber C, Cormier-Daire V. Huber C, et al. Am J Med Genet C Semin Med Genet. 2012 Aug 15;160C(3):165-74. doi: 10.1002/ajmg.c.31336. Epub 2012 Jul 12. Am J Med Genet C Semin Med Genet. 2012. PMID: 22791528 Review.
Cited by
- Whole-exome sequencing identified two novel mutations of DYNC2LI1 in fetal skeletal ciliopathy.
Zhang X, You Y, Xie X, Xu H, Zhou H, Lei Y, Sun P, Meng Y, Wang L, Lu Y. Zhang X, et al. Mol Genet Genomic Med. 2020 Dec;8(12):e1524. doi: 10.1002/mgg3.1524. Epub 2020 Oct 8. Mol Genet Genomic Med. 2020. PMID: 33030252 Free PMC article. - Autosomal recessive Stickler syndrome resulting from a COL9A3 mutation.
Hanson-Kahn A, Li B, Cohn DH, Nickerson DA, Bamshad MJ; University of Washington Center for Mendelian Genomics; Hudgins L. Hanson-Kahn A, et al. Am J Med Genet A. 2018 Dec;176(12):2887-2891. doi: 10.1002/ajmg.a.40647. Epub 2018 Nov 18. Am J Med Genet A. 2018. PMID: 30450842 Free PMC article. Review. - ARL13B regulates Sonic hedgehog signaling from outside primary cilia.
Gigante ED, Taylor MR, Ivanova AA, Kahn RA, Caspary T. Gigante ED, et al. Elife. 2020 Mar 4;9:e50434. doi: 10.7554/eLife.50434. Elife. 2020. PMID: 32129762 Free PMC article. - A postnatal role for embryonic myosin revealed by MYH3 mutations that alter TGFβ signaling and cause autosomal dominant spondylocarpotarsal synostosis.
Zieba J, Zhang W, Chong JX, Forlenza KN, Martin JH, Heard K, Grange DK, Butler MG, Kleefstra T, Lachman RS, Nickerson D, Regnier M, Cohn DH, Bamshad M, Krakow D. Zieba J, et al. Sci Rep. 2017 Feb 16;7:41803. doi: 10.1038/srep41803. Sci Rep. 2017. PMID: 28205584 Free PMC article. - Primary Cilia and Mammalian Hedgehog Signaling.
Bangs F, Anderson KV. Bangs F, et al. Cold Spring Harb Perspect Biol. 2017 May 1;9(5):a028175. doi: 10.1101/cshperspect.a028175. Cold Spring Harb Perspect Biol. 2017. PMID: 27881449 Free PMC article. Review.
References
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. - PubMed
- Baujat G, et al. Asphyxiating thoracic dysplasia: clinical and molecular review of 39 families. J Med Genet. 2013;50:91–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 HG006493/HG/NHGRI NIH HHS/United States
- UL1TR000124/TR/NCATS NIH HHS/United States
- R01 HD40182/HD/NICHD NIH HHS/United States
- KL2 TR000122/TR/NCATS NIH HHS/United States
- T32 HG002536/HG/NHGRI NIH HHS/United States
- R01 HD040182/HD/NICHD NIH HHS/United States
- R01AR062651/AR/NIAMS NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- CJX1-443835-WS-29646/PHS HHS/United States
- 1U54 HG006493/HG/NHGRI NIH HHS/United States
- P30 AR057230/AR/NIAMS NIH HHS/United States
- UM1 HG006493/HG/NHGRI NIH HHS/United States
- R01 AR066124/AR/NIAMS NIH HHS/United States
- R01 AR062651/AR/NIAMS NIH HHS/United States
- R01 GM102347/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous