Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways - PubMed (original) (raw)
Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways
Tatiana V Mishanina et al. Nat Chem Biol. 2015 Jul.
Abstract
The chemical species involved in H2S signaling remain elusive despite the profound and pleiotropic physiological effects elicited by this molecule. The dominant candidate mechanism for sulfide signaling is persulfidation of target proteins. However, the relatively poor reactivity of H2S toward oxidized thiols, such as disulfides, the low concentration of disulfides in the reducing milieu of the cell and the low steady-state concentration of H2S raise questions about the plausibility of persulfide formation via reaction between an oxidized thiol and a sulfide anion or a reduced thiol and oxidized hydrogen disulfide. In contrast, sulfide oxidation pathways, considered to be primarily mechanisms for disposing of excess sulfide, generate a series of reactive sulfur species, including persulfides, polysulfides and thiosulfate, that could modify target proteins. We posit that sulfide oxidation pathways mediate sulfide signaling and that sulfurtransferases ensure target specificity.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures
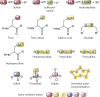
Figure 1. Structures of some biologically relevant RSS chemotypes
The use of the term ‘sulfane sulfur’ in the literature is sometimes confusing. Here, the IUPAC nomenclature for sulfane sulfur—sulfur-bonded sulfur—is used. The red (−2), green (−1), yellow (0), blue (+4), purple (+5) and orange (+6) rectangles are used to designate the valence states of sulfur as specified. In molecules containing catenated sulfurs, n ≥ 2 and R ≠ H.
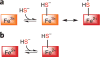
Figure 2. Models for H2S interaction with heme proteins
(a) Binding of sulfide anion to ferric heme results in the formation of ferric sulfide complex, which, depending on the polarity of the distal heme pocket, could lead to heme iron reduction and formation of the sulfur radical. Depending on the heme protein, either H2S or HS− can bind ferric heme. (b) In principle, H2S can also bind ferrous hemes, as seen with model porphyrinate complexes.
Figure 3. Potential mechanisms for persulfidation and the resolution of this modification
(a) H2S is relatively stable and is oxidized slowly by H2O2 to give HSOH. Either HSOH or HSSH formed from HSOH in the presence of a second equivalent of H2S can be attacked by a reactive cysteine on a protein to generate the persulfide modification. The bimolecular rate constants for HSOH and HSSH formation in solution at pH 7.4 and 37 °C are noted. (b) Persulfidation can result from the nucleophilic attack of a sulfide anion on an oxidized protein thiol (such as disulfide, mixed disulfide, cysteine sulfenic acid or S-nitrosylated cysteine). (c) Persulfide modifications on proteins are reversible and, unless sequestered, labile. They can be removed via persulfide interchange reactions involving glutathione (GSH), thioredoxin (Trx) or a cysteine on the same or a different protein. In all cases, the product is ultimately H2S, formed upon reduction of the persulfide moiety by either a second mole of GSH or the NADPH–thioredoxin reductase system.
Figure 4. Potential complexity associated with uncatalyzed protein modification by polysulfides
(a) A reactive thiol on a protein (dark blue sphere) could attack one of several sulfurs in a polysulfide chain generating a series of protein modifications (1–5) and eliminating H2S or polysulfides with varying number of sulfur atoms, which are omitted for clarity of presentation. This reaction complexity could be averted in an enzyme-catalyzed persulfidation reaction in which nucleophilic attack on a specific sulfur atom in the polysulfide chain is promoted. (b) Further reactions with persulfide- or polysulfide-modified proteins with a cysteine on the same or a different (light blue sphere) protein (or a small-molecule thiol) could result in homo- or heterodimerization in which the protein subunits are linked via bridging sulfur atoms. Alternatively, a vicinal cysteine residue on the modified protein could give rise to an intramolecular sulfur linkage.
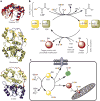
Figure 5. Rhodanese homology domain proteins and persulfide relay system for transpersulfidation
(a) Structures of three rhodanese homology domain proteins in which the active C-terminal domain is shown in red (top, S. cerevisiae putative single-domain thiosulfate sulfurtransferase (TST) YOR285, PDB 3D1P), green (middle, bovine rhodanese, PDB 1RHD) or blue (bottom, human 3-mercaptopyruvate sulfurtransferase (MST), PDB 4JGT), and the inactive N-terminal domain, when present, is shown in yellow. The active site cysteines are in ball representation, and a persulfide intermediate is seen in the rhodanese structure. (b) Sulfurtransferases such MST and TSTs (for example, rhodanese and TSTD1) and sulfide quinone oxidoreductase (SQR) accept sulfur atoms from their respective substrates (3-mercaptopyruvate for MST, thiosulfate or GSSH for TSTs and H2S for SQR) and form a persulfide intermediate. The outer sulfur can be transferred to a protein or small-molecule acceptor, resulting in a persulfide product. (c) Specificity in transpersulfidation can be achieved by enzyme-catalyzed transfer of the persulfide group. In the example shown here, either GSSH or thiosulfate transfers the outer sulfur to TST, forming a persulfide intermediate, which can be transferred to protein targets (1, 2) or to an intermediate carrier such as glutaredoxin (Grx, 3), which, in turn, transfers the persulfide to a target protein (4). Because two-cysteine-containing thiroredoxins and Grxs have vicinal resolving cysteines, the resulting persulfide modification on them is expected to be short-lived. In contrast, in carriers with a single active site cysteine, such as a subset of Grxs, the lifetime of the persulfide modification will be longer.
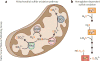
Figure 6. Sulfide oxidation pathways
(a) The canonical sulfide oxidation pathway found in most tissues resides in the mitochondrion and involves four enzymes. In the first step of the pathway, sulfide quinone oxidoreductase (SQR) oxidizes sulfide to persulfide, which is transferred from the active site of SQR to a small molecule acceptor, such as glutathione (GSH). The glutathione persulfide (GSSH) product can be oxidized by persulfide dioxygenase (PDO) to sulfite or can be used in a sulfurtransferase reaction catalyzed by rhodanese (Rhd) in the presence of sulfite, to form thiosulfate. In the final step, sulfite is oxidized to sulfate by sulfite oxidase (SO). (b) Heme-dependent sulfide oxidation pathway. An alternative pathway for sulfide oxidation involves ferric heme–dependent conversion of H2S to a mixture of thiosulfate and polysulfides. This newly discovered mechanism has been established for human hemoglobin and could be an activity of other heme proteins as well. For clarity, the fate of the H2S sulfur atom is highlighted in red and other reactants and reaction stoichiometries are omitted.
Similar articles
- Reaction of Hydrogen Sulfide with Disulfide and Sulfenic Acid to Form the Strongly Nucleophilic Persulfide.
Cuevasanta E, Lange M, Bonanata J, Coitiño EL, Ferrer-Sueta G, Filipovic MR, Alvarez B. Cuevasanta E, et al. J Biol Chem. 2015 Nov 6;290(45):26866-26880. doi: 10.1074/jbc.M115.672816. Epub 2015 Aug 12. J Biol Chem. 2015. PMID: 26269587 Free PMC article. - Synthesis, Metabolism, and Signaling Mechanisms of Hydrogen Sulfide: An Overview.
Bełtowski J. Bełtowski J. Methods Mol Biol. 2019;2007:1-8. doi: 10.1007/978-1-4939-9528-8_1. Methods Mol Biol. 2019. PMID: 31148102 Review. - Biological chemistry of hydrogen sulfide and persulfides.
Cuevasanta E, Möller MN, Alvarez B. Cuevasanta E, et al. Arch Biochem Biophys. 2017 Mar 1;617:9-25. doi: 10.1016/j.abb.2016.09.018. Epub 2016 Sep 30. Arch Biochem Biophys. 2017. PMID: 27697462 Review. - Enzymology of H2S biogenesis, decay and signaling.
Kabil O, Banerjee R. Kabil O, et al. Antioxid Redox Signal. 2014 Feb 10;20(5):770-82. doi: 10.1089/ars.2013.5339. Epub 2013 Jun 7. Antioxid Redox Signal. 2014. PMID: 23600844 Free PMC article. Review. - Kinetics of formation and reactivity of the persulfide in the one-cysteine peroxiredoxin from Mycobacterium tuberculosis.
Cuevasanta E, Reyes AM, Zeida A, Mastrogiovanni M, De Armas MI, Radi R, Alvarez B, Trujillo M. Cuevasanta E, et al. J Biol Chem. 2019 Sep 13;294(37):13593-13605. doi: 10.1074/jbc.RA119.008883. Epub 2019 Jul 16. J Biol Chem. 2019. PMID: 31311857 Free PMC article.
Cited by
- The mitochondrial NADH pool is involved in hydrogen sulfide signaling and stimulation of aerobic glycolysis.
Vitvitsky V, Kumar R, Libiad M, Maebius A, Landry AP, Banerjee R. Vitvitsky V, et al. J Biol Chem. 2021 Jan-Jun;296:100736. doi: 10.1016/j.jbc.2021.100736. Epub 2021 Apr 30. J Biol Chem. 2021. PMID: 33933447 Free PMC article. - Promoter hypomethylation and overexpression of TSTD1 mediate poor treatment response in breast cancer.
Ansar M, Thu LTA, Hung CS, Su CM, Huang MH, Liao LM, Chung YM, Lin RK. Ansar M, et al. Front Oncol. 2022 Nov 7;12:1004261. doi: 10.3389/fonc.2022.1004261. eCollection 2022. Front Oncol. 2022. PMID: 36419875 Free PMC article. - Intelligent polymeric hydrogen sulfide delivery systems for therapeutic applications.
Rong F, Wang T, Zhou Q, Peng H, Yang J, Fan Q, Li P. Rong F, et al. Bioact Mater. 2022 Apr 13;19:198-216. doi: 10.1016/j.bioactmat.2022.03.043. eCollection 2023 Jan. Bioact Mater. 2022. PMID: 35510171 Free PMC article. Review. - Structural and functional imaging of brains.
Liu Z, Zhu Y, Zhang L, Jiang W, Liu Y, Tang Q, Cai X, Li J, Wang L, Tao C, Yin X, Li X, Hou S, Jiang D, Liu K, Zhou X, Zhang H, Liu M, Fan C, Tian Y. Liu Z, et al. Sci China Chem. 2023;66(2):324-366. doi: 10.1007/s11426-022-1408-5. Epub 2022 Dec 9. Sci China Chem. 2023. PMID: 36536633 Free PMC article. Review. - β-RA reduces DMQ/CoQ ratio and rescues the encephalopathic phenotype in Coq9 R239X mice.
Hidalgo-Gutiérrez A, Barriocanal-Casado E, Bakkali M, Díaz-Casado ME, Sánchez-Maldonado L, Romero M, Sayed RK, Prehn C, Escames G, Duarte J, Acuña-Castroviejo D, López LC. Hidalgo-Gutiérrez A, et al. EMBO Mol Med. 2019 Jan;11(1):e9466. doi: 10.15252/emmm.201809466. EMBO Mol Med. 2019. PMID: 30482867 Free PMC article.
References
- Russell MJ, Hall AJ. The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J Geol Soc Lond. 1997;154:377–402. - PubMed
- Wacey D, Kilburn MR, Saunders M, Cliff J, Brasier MD. Microfossils of sulphur-metabolizing cells in 3.4-billion-year-old rocks of Western Australia. Nat Geosci. 2011;4:698–702.
- Anbar AD, Knoll AH. Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science. 2002;297:1137–1142. - PubMed
- Li C, et al. A stratified redox model for the Ediacaran ocean. Science. 2010;328:80–83. - PubMed
- Grice K, et al. Photic zone euxinia during the Permian-Triassic superanoxic event. Science. 2005;307:706–709. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources