MS2 coat proteins bound to yeast mRNAs block 5' to 3' degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system - PubMed (original) (raw)
MS2 coat proteins bound to yeast mRNAs block 5' to 3' degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system
Jennifer F Garcia et al. RNA. 2015 Aug.
Abstract
Arrays of MS2 binding sites are placed into mRNAs and are commonly used to visualize the localization of mRNAs in vivo by the expression of an MS2-GFP fusion protein. In Saccharomyces cerevisiae, we observed that arrays of MS2 binding sites inhibit 5' to 3' degradation of the mRNA in yeast cells and lead to the accumulation of a 3' mRNA fragment containing the MS2 binding sites. This accumulation can be dependent on the binding of the MS2 stem loops (MS2-SL) by MS2 coat proteins (MCPs). Since such decay fragments can still bind MCP-GFP, the localization of such mRNA fragments can complicate the interpretation of the localization of full-length mRNA in vivo.
Keywords: MS2-MCP system; mRNA decay; mRNA localization.
© 2015 Garcia and Parker; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
FIGURE 1.
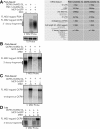
MCP binding to arrays of 24 MS2-SLs in MS2-tagged mRNAs inhibits Xrn1-mediated decay and generates 3′ mRNA decay fragments. Total mRNA was isolated and visualized by Northern using a 1.5% formaldehyde agarose gel from yeast carrying TET-off plasmids expressing either QCR8 or PGK1 mRNAs (which encodes a mitochondrial protein and a phosphoglycerate kinase, respectively) tagged with 24 MS2 stem loops (24×MS-SL) derived from pCR4-24×MS2-SL-stable plasmid (Addgene plasmid 31865) in their 3′ UTR. The white line indicates that the lanes of the gel were spliced together and reordered from the same gel. (A) Visual schematic of the MS2-tagged mRNAs analyzed in this figure that indicate the expected size of each mRNA or mRNA feature. Visualization of the MS2-tagged QCR8 or PGK1 mRNAs products expressed from a yeast Tet-off plasmid using the 3′ MS2 probe (5′-CCGCTATCGATGTTAACAGG-3′) in strains grown at 30°C to post-diauxic (PD) phase in minimal media and expressing the fusion protein, MCP-3×GFP, from the pMS2-CP-3×GFP plasmid (Haim et al. 2007). Total RNA in lane 1 was isolated from cells grown at 30°C to the PD phase in the presence of 10 µg/mL doxycycline to repress expression of the MS2-tagged QCR8 mRNA. (B) Total RNA from wild-type and xrn1Δ strains expressing the 24×MS2-tagged QCR8 mRNA and grown to PD phase were probed with 3′ MS2 probe from A. Lanes 1,2 also expressed the MCP-3×GFP fusion protein while lanes 3,4 did not. (C) Same as B but the Northern was probed with the 5′ MS2 probe (5′-CTCTACCAGCTTTGCTGTACAG-3′). (D) Total RNA from cells grown to log phase at 30°C in minimal media was probed with the 3′ MS2 probe in strains described above. To induce the expression of MCP-3×GFP during log phase from the inducible MET25 promoter, strains were grown to log phase and shifted into methionine-free minimal media for 1 h at 30°C.
FIGURE 2.
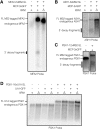
RNAs tagged with smaller arrays of MS2 binding sites also induce the production of 3′ mRNA decay fragments but not when tagged with 16 U1A stem loops. Total RNA was isolated and visualized from strains expressing mRNAs tagged with various sized MS2-SL arrays or U1A stem loops. The white line indicates that the lanes of the gel were spliced together from the same gel and reordered. Black boxes around gels indicate different gels. The MS2-tagged mRNAs are distinct from endogenous mRNAs and do not co-migrate with the 3′ mRNA decay products in A and B. (A) Total mRNA from wild-type and xrn1Δ strains expressing MFA2-2×MS-SL from pRP1083 (Sheth and Parker 2003) and MCP-3×GFP from pMS2-CP-3×GFP was run on a 6% TBE-Urea PAGE gel and visualized with a probe to the 3′ UTR of MFA2 (5′-GAT GAG AGA ATT GGA ATA AAT TAG TTT GCC AGC-3′). To induce the expression of MCP-3×GFP, strains were grown as described in Figure 1D. Lane 1 is an untagged MFA2 mRNA control. MFA2 encodes the mating pheromone, a-factor. (B) Total mRNA from wild-type and xrn1Δ strains expressing ASH1-6×MS-SL from pRJ1063 (courtesy of Ralf-Peter Jansen, Schmid et al. 2006) and MCP-YFP from pGPD-MCP-YFP was run on a 1.5% formaldehyde agarose gel and visualized with a probe to the 3′ UTR of ASH1 (5′- GTTTCGTGATAATGTCTCTTATTAGTTG-3′). To induce the expression of ASH1-6×MS2-SL mRNAs, strains were grown to log phase in minimal media containing 2% glucose and then shifted into minimal media containing 2% galactose and 1% sucrose for an hour prior to harvesting. Lane 1 is untagged Ash1 mRNA control grown under the same conditions as in A. The ASH1 mRNA encodes a component of a histone deacetylase. (C) yMK2034 (courtesy of Mark Ashe, Simpson et al. 2014) which contains an endogenously 12×MS2-SL tagged PGK1 construct was grown to as described in Figure 1D. Total mRNA was isolated from the resulting cell pellets and visualized by Northern blotting using a probe to 3′ UTR of PGK1 (5′-GAAAGAGAAAAGAAAAAAATTGATCTATCGATTTCAATTCAATTC-3′) after running on a 1.5% formaldehyde agarose gel. Lane 1 is an untagged PGK1 mRNA control. (D) Total mRNA from wild-type and xrn1Δ strains expressing PGK1-16×U1A-SL from pPS2037 (courtesy of Pamela Silver, Brodsky and Silver 2000) and U1A-GFP from pRP1194 (Brengues et al. 2005) was run on a 1.5% formaldehyde agarose gel and visualized with a probe to the 3′ UTR of PGK1 used in C.
Similar articles
- Stem-loop RNA labeling can affect nuclear and cytoplasmic mRNA processing.
Heinrich S, Sidler CL, Azzalin CM, Weis K. Heinrich S, et al. RNA. 2017 Feb;23(2):134-141. doi: 10.1261/rna.057786.116. Epub 2016 Nov 10. RNA. 2017. PMID: 28096443 Free PMC article. - Ubiquitous accumulation of 3' mRNA decay fragments in Saccharomyces cerevisiae mRNAs with chromosomally integrated MS2 arrays.
Garcia JF, Parker R. Garcia JF, et al. RNA. 2016 May;22(5):657-9. doi: 10.1261/rna.056325.116. RNA. 2016. PMID: 27090788 Free PMC article. - Using fluorescent proteins to study mRNA trafficking in living cells.
Querido E, Chartrand P. Querido E, et al. Methods Cell Biol. 2008;85:273-92. doi: 10.1016/S0091-679X(08)85012-1. Methods Cell Biol. 2008. PMID: 18155467 Review. - RNA voyeurism: A coming of age story.
Lampasona AA, Czaplinski K. Lampasona AA, et al. Methods. 2016 Apr 1;98:10-17. doi: 10.1016/j.ymeth.2015.11.024. Epub 2015 Nov 27. Methods. 2016. PMID: 26638774 Review.
Cited by
- Real-time single-molecule imaging of transcriptional regulatory networks in living cells.
Hwang DW, Maekiniemi A, Singer RH, Sato H. Hwang DW, et al. Nat Rev Genet. 2024 Apr;25(4):272-285. doi: 10.1038/s41576-023-00684-9. Epub 2024 Jan 9. Nat Rev Genet. 2024. PMID: 38195868 Review. - Real-Time Messenger RNA Dynamics in Bacillus subtilis.
Sattler L, Graumann PL. Sattler L, et al. Front Microbiol. 2021 Nov 18;12:760857. doi: 10.3389/fmicb.2021.760857. eCollection 2021. Front Microbiol. 2021. PMID: 34867890 Free PMC article. - Subcellular Transcriptomics and Proteomics: A Comparative Methods Review.
Christopher JA, Geladaki A, Dawson CS, Vennard OL, Lilley KS. Christopher JA, et al. Mol Cell Proteomics. 2022 Feb;21(2):100186. doi: 10.1016/j.mcpro.2021.100186. Epub 2021 Dec 16. Mol Cell Proteomics. 2022. PMID: 34922010 Free PMC article. Review. - A bright FIT-PNA hybridization probe for the hybridization state specific analysis of a C → U RNA edit via FRET in a binary system.
Fang GM, Chamiolo J, Kankowski S, Hövelmann F, Friedrich D, Löwer A, Meier JC, Seitz O. Fang GM, et al. Chem Sci. 2018 May 2;9(21):4794-4800. doi: 10.1039/c8sc00457a. eCollection 2018 Jun 7. Chem Sci. 2018. PMID: 29910930 Free PMC article. - In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p.
Smith C, Lari A, Derrer CP, Ouwehand A, Rossouw A, Huisman M, Dange T, Hopman M, Joseph A, Zenklusen D, Weis K, Grunwald D, Montpetit B. Smith C, et al. J Cell Biol. 2015 Dec 21;211(6):1121-30. doi: 10.1083/jcb.201503135. J Cell Biol. 2015. PMID: 26694837 Free PMC article.
References
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563. - PubMed
- Haim L, Zipor G, Aronov S, Gerst JE. 2007. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods 4: 409–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM045443/GM/NIGMS NIH HHS/United States
- R01 GM045443/GM/NIGMS NIH HHS/United States
- F32 GM108075-01A1/GM/NIGMS NIH HHS/United States
- Howard Hughes Medical Institute/United States
- F32 GM108075/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous