Pseudomonas aeruginosa high-level resistance to polymyxins and other antimicrobial peptides requires cprA, a gene that is disrupted in the PAO1 strain - PubMed (original) (raw)
Pseudomonas aeruginosa high-level resistance to polymyxins and other antimicrobial peptides requires cprA, a gene that is disrupted in the PAO1 strain
Alina D Gutu et al. Antimicrob Agents Chemother. 2015 Sep.
Abstract
The arn locus, found in many Gram-negative bacterial pathogens, mediates resistance to polymyxins and other cationic antimicrobial peptides through 4-amino-l-arabinose modification of the lipid A moiety of lipopolysaccharide. In Pseudomonas aeruginosa, several two-component regulatory systems (TCSs) control the arn locus, which is necessary but not sufficient for these resistance phenotypes. A previous transposon mutagenesis screen to identify additional polymyxin resistance genes that these systems regulate implicated an open reading frame designated PA1559 in the genome of the P. aeruginosa PAO1 strain. Resequencing of this chromosomal region and bioinformatics analysis for a variety of P. aeruginosa strains revealed that in the sequenced PAO1 strain, a guanine deletion at the end of PA1559 results in a frameshift and truncation of a full-length open reading frame that also encompasses PA1560 in non-PAO1 strains, such as P. aeruginosa PAK. Deletion analysis in the PAK strain showed that this full-length open reading frame, designated cprA, is necessary for polymyxin resistance conferred by activating mutations in the PhoPQ, PmrAB, and CprRS TCSs. The cprA gene was also required for PmrAB-mediated resistance to other cationic antimicrobial peptides in the PAK strain. Repair of the mutated cprA allele in the PAO1 strain restored polymyxin resistance conferred by an activating TCS mutation. The deletion of cprA did not affect the arn-mediated lipid A modification, indicating that the CprA protein is necessary for a different aspect of polymyxin resistance. This protein has a domain structure with a strong similarity to the extended short-chain dehydrogenase/reductase family that comprises isomerases, lyases, and oxidoreductases. These results suggest a new avenue through which to pursue targeted inhibition of polymyxin resistance.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
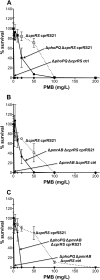
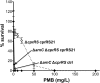
FIG 1
Effect of phoPQ, pmrAB, or phoPQ and pmrAB deletion on Pm resistance in a PAK Δ_cprRS_ cprRS21 background. For this and subsequent figures, Pm resistance experiments were performed twice; if discrepancies were seen, the experiment was performed a third time. Each panel shows a representative experiment, with the results expressed as means from three technical replicates. The error bars represent the standard deviations (SD). The nonitalicized allele name indicates the presence of an episomal version, i.e., an inducible expression strain (ctrl, empty vector control). Results from a PMB plate assay of strains induced with 0.1%
l
-Ara for 24 h are shown for strain 4537 (Δ_phoPQ_ Δ_cprRS_ cprRS21) with strain 4240 (Δ_cprRS_ cprRS21) as a positive control and strain 4541 (Δ_phoPQ_ Δ_cprRS_ ctrl) as a negative control (A), strain 4472 (Δ_pmrAB_ Δ_cprRS_ cprRS21) with strain 4240 (Δ_cprRS_ cprRS21) as a positive control and strain 4470 (Δ_pmrAB_ Δ_cprRS_ ctrl) as a negative control (B), and strain 4543 (Δ_phoPQ_ Δ_pmrAB_ Δ_cprRS_ cprRS21) with strain 4240 (Δ_cprRS_ cprRS21) as a positive control and strain 4547 (Δ_phoPQ_ Δ_pmrAB_ Δ_cprRS_ ctrl) as a negative control (C).
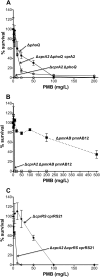
FIG 2
Effect of cprA2 deletion on Pm resistance in various PAK strain backgrounds. Shown are the results of a PMB plate assay of strain 4374 (Δ_cprA2_ Δ_phoQ_) and strain 4614 (Δ_cprA2_ Δ_phoQ_ cprA2) induced with 0.1%
l
-Ara for 24 h, with strain 2326 (Δ_phoQ_) as a positive control (A), strain 4519 (Δ_cprA2_ Δ_pmrAB_ pmrAB12) induced with 0.1%
l
-Ara for 24 h, with strain 2735 (Δ_pmrAB_ pmrAB12) as a positive control (B), and strain 4425 (Δ_cprA2_ Δ_cprRS_ cprRS21) induced with 0.1%
l
-Ara for 24 h, with strain 4240 (Δ_cprRS_ cprRS21) as a positive control (C). Empty vector control strains 4497 (Δ_cprA2_ Δ_phoQ_ pJN105), 4521 (Δ_cprA2_ Δ_pmrAB_ pJN105), and 4447 (Δ_cprA2_ Δ_cprRS_ pJN105) had Pm susceptibility values similar to those of the corresponding parental strains (not shown).
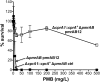
FIG 3
Repair of the cprA1 mutant allele promotes high-level Pm resistance in the PAO1 Δ_pmrAB_ strain expressing pmrAB12 in trans. Shown are the results of a PMB plate assay for strain 4763 (Δ_cprA1_::cprA + Δ_pmrAB_ pmrAB12) induced with 0.1%
l
-Ara for 24 h, with strains 4734 (Δ_pmrAB_ pmrAB12) and 4765 (Δ_cprA1_::cprA + Δ_pmrAB_ ctrl) as negative controls.
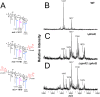
FIG 4
Deletion of cprA2 does not affect
l
-Ara4N modification of lipid A from the PAK strain. (A) Structure of lipid A with baseline hexa-acylation (m/z, 1,617), single
l
-Ara4N addition (m/z, 1,748), and double
l
-Ara4N addition (m/z, 1,879). Also shown are the MALDI-TOF spectra of lipid A isolated from strain 1026 (WT) (B), strain 2326 (Δ_phoQ_) (C), and strain 4374 (Δ_cprA2_ Δ_phoQ_) (D).
FIG 5
Effect of arnC deletion in the PAK Δ_cprRS_ cprRS21 strain. Shown are the results of a PMB plate assay of strain 4581 (Δ_arnC_ Δ_cprRS_ cprRS21) induced with 0.1%
l
-Ara for 24 h, with strain 4240 (Δ_cprRS_ cprRS21) as a positive control and strain 4586 (Δ_arnC_ Δ_cprRS_ ctrl) as a negative control.
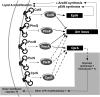
FIG 6
Two-component regulatory systems mediating resistance to Pm and other CAPs modulate expression of arn and eptA, loci that enable specific lipid A modifications, and cprA, a locus potentially implicated in other LPS modification(s) and vesiculation from the outer membrane.
Similar articles
- Polymyxin resistance of Pseudomonas aeruginosa phoQ mutants is dependent on additional two-component regulatory systems.
Gutu AD, Sgambati N, Strasbourger P, Brannon MK, Jacobs MA, Haugen E, Kaul RK, Johansen HK, Høiby N, Moskowitz SM. Gutu AD, et al. Antimicrob Agents Chemother. 2013 May;57(5):2204-15. doi: 10.1128/AAC.02353-12. Epub 2013 Mar 4. Antimicrob Agents Chemother. 2013. PMID: 23459479 Free PMC article. - PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients.
Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Høiby N, Moskowitz SM. Miller AK, et al. Antimicrob Agents Chemother. 2011 Dec;55(12):5761-9. doi: 10.1128/AAC.05391-11. Epub 2011 Oct 3. Antimicrob Agents Chemother. 2011. PMID: 21968359 Free PMC article. - PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients.
Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, Høiby N. Moskowitz SM, et al. Antimicrob Agents Chemother. 2012 Feb;56(2):1019-30. doi: 10.1128/AAC.05829-11. Epub 2011 Nov 21. Antimicrob Agents Chemother. 2012. PMID: 22106224 Free PMC article. - Adaptive resistance to cationic compounds in Pseudomonas aeruginosa.
Skiada A, Markogiannakis A, Plachouras D, Daikos GL. Skiada A, et al. Int J Antimicrob Agents. 2011 Mar;37(3):187-93. doi: 10.1016/j.ijantimicag.2010.11.019. Epub 2011 Feb 4. Int J Antimicrob Agents. 2011. PMID: 21295448 Review. - Regulating polymyxin resistance in Gram-negative bacteria: roles of two-component systems PhoPQ and PmrAB.
Huang J, Li C, Song J, Velkov T, Wang L, Zhu Y, Li J. Huang J, et al. Future Microbiol. 2020 Apr;15(6):445-459. doi: 10.2217/fmb-2019-0322. Epub 2020 Apr 6. Future Microbiol. 2020. PMID: 32250173 Free PMC article. Review.
Cited by
- Transcriptome Analysis of Pseudomonas aeruginosa Biofilm Infection in an Ex Vivo Pig Model of the Cystic Fibrosis Lung.
Harrington NE, Littler JL, Harrison F. Harrington NE, et al. Appl Environ Microbiol. 2022 Feb 8;88(3):e0178921. doi: 10.1128/AEM.01789-21. Epub 2021 Dec 8. Appl Environ Microbiol. 2022. PMID: 34878811 Free PMC article. - The Cysteine Desulfurase IscS Is a Significant Target of 2-Aminoacrylate Damage in Pseudomonas aeruginosa.
Fulton RL, Irons J, Downs DM. Fulton RL, et al. mBio. 2022 Jun 28;13(3):e0107122. doi: 10.1128/mbio.01071-22. Epub 2022 Jun 2. mBio. 2022. PMID: 35652590 Free PMC article. - Evolutionary trade-offs associated with loss of PmrB function in host-adapted Pseudomonas aeruginosa.
Bricio-Moreno L, Sheridan VH, Goodhead I, Armstrong S, Wong JKL, Waters EM, Sarsby J, Panagiotou S, Dunn J, Chakraborty A, Fang Y, Griswold KE, Winstanley C, Fothergill JL, Kadioglu A, Neill DR. Bricio-Moreno L, et al. Nat Commun. 2018 Jul 6;9(1):2635. doi: 10.1038/s41467-018-04996-x. Nat Commun. 2018. PMID: 29980663 Free PMC article. - The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions.
Jochumsen N, Marvig RL, Damkiær S, Jensen RL, Paulander W, Molin S, Jelsbak L, Folkesson A. Jochumsen N, et al. Nat Commun. 2016 Oct 3;7:13002. doi: 10.1038/ncomms13002. Nat Commun. 2016. PMID: 27694971 Free PMC article. - In-depth biological characterization of two black soldier fly anti-Pseudomonas peptides reveals LPS-binding and immunomodulating effects.
Van Moll L, Wouters M, De Smet J, De Vooght L, Delputte P, Van Der Borght M, Cos P. Van Moll L, et al. mSphere. 2023 Oct 24;8(5):e0045423. doi: 10.1128/msphere.00454-23. Epub 2023 Oct 6. mSphere. 2023. PMID: 37800918 Free PMC article.
References
- Moskowitz SM, Silva SJ, Mayer-Hamblett N, Pasta DJ, Mink DR, Mabie JA, Konstan MW, Wagener JS, Investigators and Coordinators of the Epidemiologic Study of Cystic Fibrosis (ESCF). 2008. Shifting patterns of inhaled antibiotic use in cystic fibrosis. Pediatr Pulmonol 43:874–881. doi:10.1002/ppul.20873. - DOI - PubMed
- Alonso A, Campanario E, Martinez JL. 1999. Emergence of multidrug-resistant mutants is increased under antibiotic selective pressure in Pseudomonas aeruginosa. Microbiology 145(Pt 10):2857–2862. - PubMed
- Foweraker JE, Laughton CR, Brown DF, Bilton D. 2009. Comparison of methods to test antibiotic combinations against heterogeneous populations of multiresistant Pseudomonas aeruginosa from patients with acute infective exacerbations in cystic fibrosis. Antimicrob Agents Chemother 53:4809–4815. doi:10.1128/AAC.00269-09. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical