An Alternative Phosphorylation Switch in Integrin β2 (CD18) Tail for Dok1 Binding - PubMed (original) (raw)
An Alternative Phosphorylation Switch in Integrin β2 (CD18) Tail for Dok1 Binding
Sebanti Gupta et al. Sci Rep. 2015.
Abstract
Integrins are involved in cell migration and adhesion. A large number of proteins interact with the cytoplasmic tails of integrins. Dok1 is a negative regulator of integrin activation and it binds to the phosphorylated membrane proximal NxxY motif in a number of integrin β tails. The β tail of the β2 integrins contains a non-phosphorylatable NxxF motif. Hence it is unclear how Dok1 associates with the β2 integrins. We showed in this study using NMR and cell based analyses that residues Ser745 and Ser756 in the integrin β2 tail, which are adjacent to the NxxF motif, are required for Dok1 interaction. NMR analyses detected significant chemical shift changes and higher affinity interactions between Dok1 phospho-tyrosine binding (PTB) domain and integrin β2 tail peptide containing pSer756 compared to pSer745. The phosphorylated β2 peptide occupies the canonical ligand binding pocket of Dok1 based on the docked structure of the β2 tail-Dok1 PTB complex. Taken together, our data suggest an alternate phosphorylation switch in β2 integrins that regulates Dok1 binding. This could be important for cells of the immune system and their functions.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
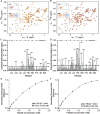
Figure 1. Dok1 binds to the integrin β2 tail upon phosphorylation of Ser745 and Ser756.
(A and B) represent the 15N-1H HSQC spectra of the Dok1 PTB domain, showing chemical shift changes at 0 mM (red contour), 0.4 mM (cyan contour), 1 mM (violet contour) and 2 mM (orange contour) concentrations of pSer745 (A) and pSer756 β2 tail (B). (in inset) Chemical shift changes of residue Arg69 and Val61 of the PTB domain of Dok1 are shown. Bar diagrams showing combined chemical shift perturbation for 15N and HN resonances of each residue of the Dok1 PTB domain upon binding to pSer745-β2 (C) and pSer756-β2 (D). Note in (D) residues showing resonance broadening upon additions of phosphorylated β2 tail are marked as asterisks. The dotted lines in (C and D) marked average chemical shift perturbation. The secondary structural elements are shown at the top of each plot. Normalized chemical shift differences are plotted against the concentrations of pSer745-β2 (E) and pSer756-β2 (F) for Dok1 PTB domain to determine equilibrium dissociation constants (Kd) values.
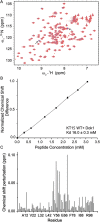
Figure 2. Interactions of Dok1 with unphosphorylated β2 tail KT15 peptide.
(A) Overlay of 15N-1H HSQC spectra of the Dok1 PTB domain, showing chemical shift changes at protein:peptide concentration ratios 1:0 (red contour), 1:1 (cyan contour), 1:3 (violet contour) and 1:6 (orange contour). Two HSQC peaks corresponding to residue Arg54 at ~9.00 ppm and residue Tyr56 at ~7.9 ppm showed KT15 binding induced perturbation (arrows). (B) Normalized chemical shift differences for Dok1 PTB domain are plotted against the concentrations of KT15 peptide to determine the Kd value. (C) Bar diagram showing combined chemical shift perturbations for 15N and HN resonances of each residue of the Dok1 PTB domain upon binding to KT15 peptide (protein:peptide concentration ratio 1:6).
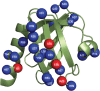
Figure 3. Mapping residues of Dok1 PTB domain that interact with pSer756-β2 tail.
A ribbon representation of the three-dimensional structure of the Dok1 PTB domain (pdb: 2v76). Residues that exhibited above average chemical shift perturbations and resonance broadening in the presence of pSer756-β2 tail are shown as blue spheres and red spheres, respectively.
Figure 4. Analyses of interactions of pSer756 KT15 peptide with Dok1 PTB domain.
(A) Bar diagram showing combined chemical shift perturbations for 15N and HN resonances of residues of the Dok1 PTB domain upon binding to pSer756 KT15 peptide. Residues showing resonance broadening are marked (asterisks). (B) Normalized chemical shift differences for Dok1 PTB domain are plotted against the concentrations of pSer756 KT15 peptide to determine the Kd value. (C) 31P NMR spectra of pSer756 KT15 peptide in free solution and in the presence of Dok1 PTB domain at different peptide:protein molar ratios (1:0 (red), 1:0.5 (blue), 1:1 (cyan) and 1:3 (green)). (D) Saturation transfer difference (STD) NMR spectrum of pSer756-KT15 peptide in the presence of Dok1 PTB domain. The off-resonance or the reference spectrum is shown (top).
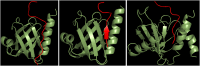
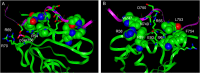
Figure 5. Binding mode and orientation of pSer756 KT15 peptide in complex with Dok1 PTB domain.
(left panel) A ribbon representation of the overall topology of the Dok1 PTB domain (green) in complex with pSer756-KT15 peptide (red) obtained from docking simulation. (middle panel) X-ray structure of Dok1 PTB domain (green) in complex with Tyr phosphorylated RET peptide (red). (right panel) NMR structure of the Numb PTB domain (green) in complex with Nak peptide (red).
Figure 6. Molecular interactions of pSer756 KT15 peptide with Dok1 PTB domain.
(A) The docked model of pSer756-KT15/Dok1 PTB domain suggests potential salt bridge/hydrogen bond interactions between the phosphate group of pSer756 and the side chain guanidinium groups of Arg54, Arg69 and Arg70 of Dok1. (B) The probable aromatic-aromatic stacking and cation-π interactions of Dok1 PTB domain Phe89 and Arg58 with Trp747 of pSer756-KT15 peptide are shown. Hydrophobic packing interactions between Phe754 and Leu753 in the NPLF motif of pSer756-KT15 peptide with Ile96 of the PTB domain are observed. In addition, there can be polar and ionic interactions: Asp749 (K15) with Glu93 (Dok1) and Asp750 (K15) with Arg55 (Dok1).
Figure 7. Analyses of the interaction between integrin β2 tail and Dok1 in cells.
(A) K562 cells transfected with the indicated expression plasmids were lysed. Proteins were resolved on 10% SDS-PAGE under reducing conditions and immunoblotted (IB) with either anti-GFP or anti-Dok1 antibodies. (B) Transfected K562 cells were subjected to YFP-photobleach FRET analyses. Each data point represents the mean ± S.D. of ≥30 cells analyzed. (C) Flow cytometry analyses of K562 stable line expressing Dok1-CFP that were transfected with integrin αLβ2-YFP and CCR5. Wild-type K562 cells were used as the control group. (D) FRET analyses of cells in (C) that were treated without or with chemokine RANTES (50 ng/ml) for 10 min at 37 °C. 60 and 39 cells were analyzed for conditions without and with RANTES treatment, respectively. Data point represent mean ± S.D. *,p < 0.05, Student’s t test.
Similar articles
- Leukocyte arrest: Biomechanics and molecular mechanisms of β2 integrin activation.
Fan Z, Ley K. Fan Z, et al. Biorheology. 2015;52(5-6):353-77. doi: 10.3233/BIR-15085. Biorheology. 2015. PMID: 26684674 Free PMC article. Review. - Interaction Analyses of 14-3-3ζ, Dok1, and Phosphorylated Integrin β Cytoplasmic Tails Reveal a Bi-molecular Switch in Integrin Regulation.
Chatterjee D, D'Souza A, Zhang Y, Bin W, Tan SM, Bhattacharjya S. Chatterjee D, et al. J Mol Biol. 2018 Oct 19;430(21):4419-4430. doi: 10.1016/j.jmb.2018.09.008. Epub 2018 Sep 20. J Mol Biol. 2018. PMID: 30243836 - An integrin phosphorylation switch: the effect of beta3 integrin tail phosphorylation on Dok1 and talin binding.
Oxley CL, Anthis NJ, Lowe ED, Vakonakis I, Campbell ID, Wegener KL. Oxley CL, et al. J Biol Chem. 2008 Feb 29;283(9):5420-6. doi: 10.1074/jbc.M709435200. Epub 2007 Dec 21. J Biol Chem. 2008. PMID: 18156175 - Beta integrin tyrosine phosphorylation is a conserved mechanism for regulating talin-induced integrin activation.
Anthis NJ, Haling JR, Oxley CL, Memo M, Wegener KL, Lim CJ, Ginsberg MH, Campbell ID. Anthis NJ, et al. J Biol Chem. 2009 Dec 25;284(52):36700-36710. doi: 10.1074/jbc.M109.061275. Epub 2009 Oct 20. J Biol Chem. 2009. PMID: 19843520 Free PMC article. - Intracellular β2 integrin (CD11/CD18) interacting partners in neutrophil trafficking.
Thome S, Begandt D, Pick R, Salvermoser M, Walzog B. Thome S, et al. Eur J Clin Invest. 2018 Nov;48 Suppl 2:e12966. doi: 10.1111/eci.12966. Epub 2018 Jun 29. Eur J Clin Invest. 2018. PMID: 29896791 Review.
Cited by
- How integrin phosphorylations regulate cell adhesion and signaling.
Gahmberg CG, Grönholm M. Gahmberg CG, et al. Trends Biochem Sci. 2022 Mar;47(3):265-278. doi: 10.1016/j.tibs.2021.11.003. Epub 2021 Dec 4. Trends Biochem Sci. 2022. PMID: 34872819 Free PMC article. Review. - Leukocyte arrest: Biomechanics and molecular mechanisms of β2 integrin activation.
Fan Z, Ley K. Fan Z, et al. Biorheology. 2015;52(5-6):353-77. doi: 10.3233/BIR-15085. Biorheology. 2015. PMID: 26684674 Free PMC article. Review. - The structural basis of β2 integrin intra-cellular multi-protein complexes.
Bhattacharjya S. Bhattacharjya S. Biophys Rev. 2022 Sep 7;14(5):1183-1195. doi: 10.1007/s12551-022-00995-x. eCollection 2022 Oct. Biophys Rev. 2022. PMID: 36345283 Free PMC article. Review. - LFA-1 Activation in T-Cell Migration and Immunological Synapse Formation.
Shi H, Shao B. Shi H, et al. Cells. 2023 Apr 12;12(8):1136. doi: 10.3390/cells12081136. Cells. 2023. PMID: 37190045 Free PMC article. Review. - Post-translational modification-regulated leukocyte adhesion and migration.
Loh JT, Su IH. Loh JT, et al. Oncotarget. 2016 Jun 14;7(24):37347-37360. doi: 10.18632/oncotarget.8135. Oncotarget. 2016. PMID: 26993608 Free PMC article. Review.
References
- Hynes R. O. Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 (2002). - PubMed
- Tan S. M. The leucocyte β2 (CD18) integrins: the structure, functional regulation and signalling properties. Bioscience Reports 32, 241–269 (2012) - PubMed
- Tadokoro S. et al. Talin binding to integrin beta tails: a final common step in integrin activation. Science 302, 103–106 (2003). - PubMed
- Cambell I. D. & Ginsberg M. H. The talin-tail interaction places integrin activation on FERM ground. Trends Biochem. Sci. 29, 429–435 (2004). - PubMed
- Senetar M. A. & McCann R. O. Gene duplication and functional divergence during evolution of the cytoskeletal linker protein talin. Gene 362, 141–152 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources