Identification and characterization of a HEPN-MNT family type II toxin-antitoxin in Shewanella oneidensis - PubMed (original) (raw)
Identification and characterization of a HEPN-MNT family type II toxin-antitoxin in Shewanella oneidensis
Jianyun Yao et al. Microb Biotechnol. 2015 Nov.
Abstract
Toxin-antitoxin (TA) systems are prevalent in bacteria and archaea. However, related studies in the ecologically and bioelectrochemically important strain Shewanella oneidensis are limited. Here, we show that SO_3166, a member of the higher eukaryotes and prokaryotes nucleotide-binding (HEPN) superfamily, strongly inhibited cell growth in S. oneidensis and Escherichia coli. SO_3165, a putative minimal nucleotidyltransferase (MNT), neutralized the toxicity of SO_3166. Gene SO_3165 lies upstream of SO_3166, and they are co-transcribed. Moreover, the SO_3165 and SO_3166 proteins interact with each other directly in vivo, and antitoxin SO_3165 bound to the promoter of the TA operon and repressed its activity. Finally, the conserved Rx4-6H domain in HEPN family was identified in SO_3166. Mutating either the R or H abolished SO_3166 toxicity, confirming that Rx4-6H domain is critical for SO_3166 activity. Taken together, these results demonstrate that SO_3166 and SO_3165 in S. oneidensis form a typical type II TA pair. This TA pair plays a critical role in regulating bacterial functions because its disruption led to impaired cell motility in S. oneidensis. Thus, we demonstrated for the first time that HEPN-MNT can function as a TA system, thereby providing important insights into the understanding of the function and regulation of HEPNs and MNTs in prokaryotes.
© 2015 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures
Figure 1
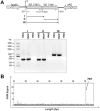
SO_3166 is the toxin and SO_3165 is the antitoxin. E . coli K12 BW25113 hosts containing pCA24N-based constructs were cultured in LB medium supplemented with 30 μg/μl chloramphenicol and 1 mM IPTG (added at OD600 of 0.1). Cell growth (A) and viability (CFU/mL) (B) were tested at the time points indicated. The cells induced for 5 h were serially diluted, dropped onto LB plates and incubated at 37°C for 16 hr (C). MR-1 hosts carrying the pHGE-based plasmids were cultured in LB with 50 μg/mL kanamycin, and 1 mM IPTG were added at OD600 ∼ 0.1. Cell growth (D) and viability (E) were tested at the time points indicated. (F) MR-1 hosts carrying the pHGE-based plasmids were streaked onto LB plates with 50 μg/mL kanamycin with or without 1 mM IPTG, and were incubated for 16 hr. Data are from two independent cultures, and standard deviations are shown in A, B, D and E.
Figure 2
Co-transcription of SO _3165 and SO _3166. (A) Primers were designed to amplify the whole open reading frame of SO_3165 (I), SO_3166 (II) and the total region that covers the start codon of SO_3165 and stop codon of SO_3166 (III). Approximately 150 ng of cDNA reverse transcribed from MR-1 RNA was used as templates to amplify the three fragments. The same amount of MR-1 genomic DNA and RNA were used as the positive and negative controls, respectively. M indicates the DNA ladder. (B) Primer extension was conducted using the 5′ FAM-labeled reverse transcriptional primer FAM-SO_3166-r and total RNA isolated from E. coli K12 BW25113 carrying the pBS(Kan)-SO_3165-3166 plasmid. The X-axis indicates the length of the cDNA with FAM and the Y-axis indicates intensity of the fluorescence signal.
Figure 3
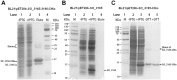
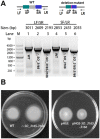
SO_3165 and SO_3166 form a complex in vivo. (A) Toxin SO_3166 with a C-terminal hexahistidine tag (His-tagged) was constructed together with untagged antitoxin SO_3165 to generate pET28b-SO __3165-3166_-CHis. After induction with 1 mM IPTG, the 16.00 kDa SO_3166-CHis and a 15.57 kDa SO_3165 were induced (lane 2). The negative control was included when no IPTG was added (lane 1). During purification, SO_3166-CHis and SO_3165 were co-purified (lane 3). The protein marker (M) was loaded in lane 4. (B) SO_3165 was induced and purified via pET28b-SO _3165 with IPTG induction under the same condition described in (A). The purified SO_3165 cannot bind to the Ni-NTA agarose beads (lane 4). (C). SO_3165-CHis (16.39 kDa) was induced and purified via pET28b-SO __3165_-CHis with IPTG induction followed the same condition described in (A). Dimerization of SO_316 was observed (lane 4), and the addition of the reducing agent dithiothreitol (DTT) greatly reduced the dimerization (lane 5).
Figure 4
Antitoxin SO_3165 binds to the promoter of the SO _3165–3166 operon. (A) The sequence of the promoter DNA used for EMSA (296-nt upstream of the translational start of the operon). The double underlines indicated the primers used for PCR amplification for the promoter region. The palindromic sequences are highlighted in blue, while the sequence between the two arms are highlighted in violet. (B) EMSA results demonstrating that purified SO_3165-CHis binds to the 296 bp biotin-labeled promoter DNA of SO _3165. The binding increases with the increasing concentrations of SO_3165-CHis protein. (C) Mid-log-phase cells of the indicated strains carrying the integrated reporter system (300-nt upstream of the translational start of the operon) were collected and tested for β-galactosidase activity. Error bars represent standard deviations for triplicate cultures. Asterisks represent a statistically significant difference between the wild-type and indicated mutants (P < 0.001; n = 3).
Figure 5
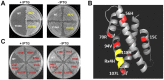
Key residues for determining SO_3166 toxicity. (A) Toxicity results of single-site mutagenesis of the R and H in Rx4H region and an adjacent tyrosine of toxin SO_3166 in the pCA24N-SO _3166 plasmid in DH5α. (B) Predicted 3-D structure of SO_3166. The conserved domain Rx4H is situated at the end of one helix (yellow). Other residues obtained by epPCR assay that affected SO_3166 toxicity are shown in red. (C) Toxicity test of seven strains expressing different mutated SO_3166 proteins obtained by epPCR. WT indicates the wild-type SO_3166 protein; the remainders are mutated proteins. The number in the mutated protein indicates the position of the amino acid in SO_3166. Overnight cultures were streaked on 30 μg_/_mL chloramphenicol LB plates with or without 0.5 mM IPTG. Two independent cultures were evaluated for each; only one representative image is shown here.
Figure 6
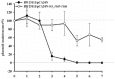
SO_3166-SO_3165 confers plasmid stability in E. coli. E . coli K12 BW25113 harboring plasmids pCA24N and pCA24N-SO _3165-3166 were used for the plasmid stability assay. Overnight cultures were 1% diluted in LB medium without any antibiotics, then incubated at 37°C for 12 hr. This process was repeated every 12 hr for 7 days. Three independent cultures were conducted, and the data are shown as means ± standard deviations.
Figure 7
SO_3166-SO_3165 represses motility in S. oneidensis. (A) In-frame deletion mutant of the toxin gene SO _3166 and the SO _3165- SO _3166 operon. Primer LF indicates the long forward primer and SF indicates the short forward primer. Primers SR and LR indicate the short reverse and long reverse primers, respectively. PCR products are indicated with the expected sizes using genomic DNA from the wild-type (WT) and the deletion mutants. (B) Swimming motility test for the SO_ 3165-3166 deletion mutant versus WT (left plate), and for wild-type MR-1 co-expressing SO _3165 and SO _3166 versus empty vector (right plate). Two independent cultures were evaluated for each; only one representative image is shown here.
Similar articles
- Structure-function analyses reveal the molecular architecture and neutralization mechanism of a bacterial HEPN-MNT toxin-antitoxin system.
Jia X, Yao J, Gao Z, Liu G, Dong YH, Wang X, Zhang H. Jia X, et al. J Biol Chem. 2018 May 4;293(18):6812-6823. doi: 10.1074/jbc.RA118.002421. Epub 2018 Mar 19. J Biol Chem. 2018. PMID: 29555683 Free PMC article. - Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system.
Yao J, Zhen X, Tang K, Liu T, Xu X, Chen Z, Guo Y, Liu X, Wood TK, Ouyang S, Wang X. Yao J, et al. Nucleic Acids Res. 2020 Nov 4;48(19):11054-11067. doi: 10.1093/nar/gkaa855. Nucleic Acids Res. 2020. PMID: 33045733 Free PMC article. - Type II toxin/antitoxin system ParESO /CopASO stabilizes prophage CP4So in Shewanella oneidensis.
Yao J, Guo Y, Wang P, Zeng Z, Li B, Tang K, Liu X, Wang X. Yao J, et al. Environ Microbiol. 2018 Mar;20(3):1224-1239. doi: 10.1111/1462-2920.14068. Epub 2018 Mar 25. Environ Microbiol. 2018. PMID: 29411516 - Regulation of toxin-antitoxin systems by proteolysis.
Brzozowska I, Zielenkiewicz U. Brzozowska I, et al. Plasmid. 2013 Jul;70(1):33-41. doi: 10.1016/j.plasmid.2013.01.007. Epub 2013 Feb 8. Plasmid. 2013. PMID: 23396045 Review. - Toxin-antitoxin systems in bacteria and archaea.
Yamaguchi Y, Park JH, Inouye M. Yamaguchi Y, et al. Annu Rev Genet. 2011;45:61-79. doi: 10.1146/annurev-genet-110410-132412. Annu Rev Genet. 2011. PMID: 22060041 Review.
Cited by
- Toxin-antitoxin systems in bacterial pathogenesis.
Sonika S, Singh S, Mishra S, Verma S. Sonika S, et al. Heliyon. 2023 Mar 3;9(4):e14220. doi: 10.1016/j.heliyon.2023.e14220. eCollection 2023 Apr. Heliyon. 2023. PMID: 37101643 Free PMC article. Review. - Toxin-Antitoxin Gene Pairs Found in Tn_3_ Family Transposons Appear To Be an Integral Part of the Transposition Module.
Lima-Mendez G, Oliveira Alvarenga D, Ross K, Hallet B, Van Melderen L, Varani AM, Chandler M. Lima-Mendez G, et al. mBio. 2020 Mar 31;11(2):e00452-20. doi: 10.1128/mBio.00452-20. mBio. 2020. PMID: 32234815 Free PMC article. - Mobile Genetic Elements and Evolution of CRISPR-Cas Systems: All the Way There and Back.
Koonin EV, Makarova KS. Koonin EV, et al. Genome Biol Evol. 2017 Oct 1;9(10):2812-2825. doi: 10.1093/gbe/evx192. Genome Biol Evol. 2017. PMID: 28985291 Free PMC article. Review. - Characterization of the Deep-Sea Streptomyces sp. SCSIO 02999 Derived VapC/VapB Toxin-Antitoxin System in Escherichia coli.
Guo Y, Yao J, Sun C, Wen Z, Wang X. Guo Y, et al. Toxins (Basel). 2016 Jul 1;8(7):195. doi: 10.3390/toxins8070195. Toxins (Basel). 2016. PMID: 27376329 Free PMC article. - Bacterial toxin-antitoxin modules: classification, functions, and association with persistence.
Singh G, Yadav M, Ghosh C, Rathore JS. Singh G, et al. Curr Res Microb Sci. 2021 Jul 7;2:100047. doi: 10.1016/j.crmicr.2021.100047. eCollection 2021 Dec. Curr Res Microb Sci. 2021. PMID: 34841338 Free PMC article. Review.
References
- Belitsky M, Avshalom H, Erental A, Yelin I, Kumar S, London N, et al. The Escherichia coli Extracellular Death Factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol Cell. 2011;41:625–635. - PubMed
- Bernard P. Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226:735–745. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources