Structure and gating of the nuclear pore complex - PubMed (original) (raw)
Structure and gating of the nuclear pore complex
Matthias Eibauer et al. Nat Commun. 2015.
Abstract
Nuclear pore complexes (NPCs) perforate the nuclear envelope and allow the exchange of macromolecules between the nucleus and the cytoplasm. To acquire a deeper understanding of this transport mechanism, we analyse the structure of the NPC scaffold and permeability barrier, by reconstructing the Xenopus laevis oocyte NPC from native nuclear envelopes up to 20 Å resolution by cryo-electron tomography in conjunction with subtomogram averaging. In addition to resolving individual protein domains of the NPC constituents, we propose a model for the architecture of the molecular gate at its central channel. Furthermore, we compare and contrast this native NPC structure to one that exhibits reduced transport activity and unveil the spatial properties of the NPC gate.
Figures
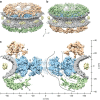
Figure 1. The structure of the native X. laevis NPC.
(a,b) Surface-rendered grazing views of the NPC. The NE is depicted in grey, the luminal densities in yellow, and the SR in blue. (a) Upper side of the CPR is shown in golden colour. (b) Top of the NPR is shown in green colour. (c) View of the central nucleocytoplasmic section (25-nm thick) through the NPC structure; CCR (A) and SR-CCR interface (B). Extended linker structures protrude from the NPR (C and D), as well as from the CPR (E). The putative position of the Nup214/Nup88 complexes is denoted by F. Suggested nuclear transport routes passing through NPC barrier, illustrated as solid and dashed curves. The axes show the dimensions of the NPC in the _x_- and _y_-direction.
Figure 2. Docking of the Y-shaped complex.
The two highest ranked docking solutions of the Y-shaped complex into the CPR electron density (in golden colour) are shown in transparent light blue for the inner position and transparent light green for the outer position. The NE is omitted for clarity, membrane connections are indicated with grey arrowheads. (a) Surface-rendered grazing view of the CPR. The putative positions of Seh1 in the inner and outer short arms are marked by white arrowheads. The suggested position of Nup37 in the inner long arm is indicated by a black arrowhead. (b) Side view of the CPR asymmetric unit. The proposed position of the Nup160 β-propeller domain in the inner long arm is labelled by a black arrowhead. In addition, a β-propeller-like structure is indicated in the outer long arm by a white arrowhead. (c) The CPR is oriented and sectioned such that it shows the electron densities of the inner Y-shaped complex from the top. The assumed positions of Seh1 and Nup37 are marked by white and black arrowheads, respectively. (d) The same for the outer Y-shaped complex. The assumed positions of Seh1 and Nup43 are marked by white and black arrowheads, respectively. Scale bar, 5 nm.
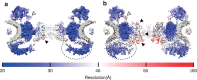
Figure 3. Structural differences in the nucleocytoplasmic section.
(a) View of the central nucleocytoplasmic section through the native NPC. (b) View of the central nucleocytoplasmic section through the ActD-NPC. Both sections are 25-nm thick. The putative Nup214 region is indicated by white arrowheads. The local resolution of the structures is visualized by surface colouring. Resolution values are given by the colour key. The dashed ellipses indicate major structural changes in the NPR. Regions in the CCR with comparatively high structural flexibility are indicated by black arrowheads.
Figure 4. Structural differences in the central channel.
(a) The central _x–y_-section (10-nm thick) through the native NPC shows the organization of the central channel; the SR and CCR are depicted in blue, the NE in grey and the luminal densities in yellow. Channels marked by orange dots (2 nm in diameter) pass through the SR-CCR interfaces at a radius of ∼23 nm with respect to the centre of the channel (suggested transport route, Fig. 1c, dashed line). (b) Due to reduced transport activity, the corresponding view of the ActD-NPC exhibits an intact outer pore ring with a diameter of ∼37 nm. In addition, the inner pore ring adopts a distinct structure (purple). (c,d) Magnified views (24 × 24 nm2) of the SR-CCR interfaces indicate their structural changes. The channel, enclosed by the connecting filaments (arrows) in the native NPC (c), is blocked in the ActD-NPC (d).
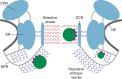
Figure 5. Schematic model of the NPC architecture and gating.
The scaffold of the NPC is depicted in cyan and the NE in dark grey. The NPR and linker structures between the ring moieties are illustrated in light grey. FG domains at the NPR construct a repulsive entropic barrier (blue wavy lines), which collapses on interaction with cargo–receptor complexes (depicted as green and purple circles, respectively). The selective phase within the central pore (red wavy lines) exhibits an ordered architecture that is structurally altered during transport.
Similar articles
- Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture.
Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Frenkiel-Krispin D, et al. J Mol Biol. 2010 Jan 22;395(3):578-86. doi: 10.1016/j.jmb.2009.11.010. Epub 2009 Nov 11. J Mol Biol. 2010. PMID: 19913035 - Insights into the gate of the nuclear pore complex.
Zwerger M, Eibauer M, Medalia O. Zwerger M, et al. Nucleus. 2016;7(1):1-7. doi: 10.1080/19491034.2015.1130197. Epub 2016 Feb 22. Nucleus. 2016. PMID: 26902931 Free PMC article. - Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport.
Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. Stoffler D, et al. J Mol Biol. 2003 Apr 18;328(1):119-30. doi: 10.1016/s0022-2836(03)00266-3. J Mol Biol. 2003. PMID: 12684002 - Nuclear pore complex structure and plasticity revealed by electron and atomic force microscopy.
Maco B, Fahrenkrog B, Huang NP, Aebi U. Maco B, et al. Methods Mol Biol. 2006;322:273-88. doi: 10.1007/978-1-59745-000-3_19. Methods Mol Biol. 2006. PMID: 16739730 Review. - The role of nuclear envelope calcium in modifying nuclear pore complex structure.
Erickson ES, Mooren OL, Moore D, Krogmeier JR, Dunn RC. Erickson ES, et al. Can J Physiol Pharmacol. 2006 Mar-Apr;84(3-4):309-18. doi: 10.1139/y05-109. Can J Physiol Pharmacol. 2006. PMID: 16902578 Review.
Cited by
- From integrative structural biology to cell biology.
Sali A. Sali A. J Biol Chem. 2021 Jan-Jun;296:100743. doi: 10.1016/j.jbc.2021.100743. Epub 2021 May 4. J Biol Chem. 2021. PMID: 33957123 Free PMC article. Review. - Mapping the native organization of the yeast nuclear pore complex using nuclear radial intensity measurements.
Vallotton P, Rajoo S, Wojtynek M, Onischenko E, Kralt A, Derrer CP, Weis K. Vallotton P, et al. Proc Natl Acad Sci U S A. 2019 Jul 16;116(29):14606-14613. doi: 10.1073/pnas.1903764116. Epub 2019 Jul 1. Proc Natl Acad Sci U S A. 2019. PMID: 31262825 Free PMC article. - Physics of the Nuclear Pore Complex: Theory, Modeling and Experiment.
Hoogenboom BW, Hough LE, Lemke EA, Lim RYH, Onck PR, Zilman A. Hoogenboom BW, et al. Phys Rep. 2021 Jul 25;921:1-53. doi: 10.1016/j.physrep.2021.03.003. Epub 2021 Mar 24. Phys Rep. 2021. PMID: 35892075 Free PMC article. - Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex.
Zhang Y, Li S, Zeng C, Huang G, Zhu X, Wang Q, Wang K, Zhou Q, Yan C, Zhang W, Yang G, Liu M, Tao Q, Lei J, Shi Y. Zhang Y, et al. Cell Res. 2020 Jun;30(6):532-540. doi: 10.1038/s41422-020-0320-y. Epub 2020 May 4. Cell Res. 2020. PMID: 32367042 Free PMC article. - Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis.
Huang G, Zhang Y, Zhu X, Zeng C, Wang Q, Zhou Q, Tao Q, Liu M, Lei J, Yan C, Shi Y. Huang G, et al. Cell Res. 2020 Jun;30(6):520-531. doi: 10.1038/s41422-020-0319-4. Epub 2020 May 6. Cell Res. 2020. PMID: 32376910 Free PMC article.
References
- Hoelz A., Debler E. W. & Blobel G. The structure of the nuclear pore complex. Annu. Rev. Biochem. 80, 613–643 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources