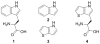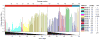Chemical Evolution of a Bacterial Proteome - PubMed (original) (raw)
Chemical Evolution of a Bacterial Proteome
Michael Georg Hoesl et al. Angew Chem Int Ed Engl. 2015.
Abstract
We have changed the amino acid set of the genetic code of Escherichia coli by evolving cultures capable of growing on the synthetic noncanonical amino acid L-β-(thieno[3,2-b]pyrrolyl)alanine ([3,2]Tpa) as a sole surrogate for the canonical amino acid L-tryptophan (Trp). A long-term cultivation experiment in defined synthetic media resulted in the evolution of cells capable of surviving Trp→[3,2]Tpa substitutions in their proteomes in response to the 20,899 TGG codons of the E. coli W3110 genome. These evolved bacteria with new-to-nature amino acid composition showed robust growth in the complete absence of Trp. Our experimental results illustrate an approach for the evolution of synthetic cells with alternative biochemical building blocks.
Keywords: Escherichia coli; continuous evolution; genetic code translation; synthetic biology; tryptophan analogues.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
Figure 1
Structures of 1: tryptophan (Trp), 2: indole (Ind), 3: β-thieno[3,2_-b_]-pyrrole ([3,2]Tp), and L-β-(thieno[3,2_-b_]pyrrolyl)alanine ([3,2]Tpa).
Figure 2. Adaptation of E. coli strain CGSC# 7679 to [3,2]Tpa
The upper scale indicates the passage count while the lower scale shows the time in days. In each passage, OD600nm was measured. The growth temperature was decreased from 37°C to 30°C while going from NMM2s to NMM1. The different medium compositions are indicated on the right side and are described in details in section 4 in the SI.
Figure 3
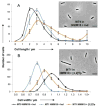
Morphology and average distribution of cell length (A) and width (B) as measured by microscopy of MT0 and MT1. Strains were grown in NMM19 with either 100 μM of Indole (black dots → MT0; blue diamonds → MT1) or of [3,2]Tp (orange squares → MT1). Error bars represent the standard errors of the means of four experiments (with 350 cells studied by experiment). Inserts: examples of brightfield microscopy pictures of MT0 (0 μM [3,2]Tp /100 μM Indole) and MT1 (0 μM Indole/100 μM [3,2]Tp) cells grown in NMM19 and prepared on a 1% agarose-coated slide to be visualised by microscopy. The calibration bar indicates 5 μm. Details can be found in sections 9 and 10 in the SI.
Similar articles
- Proteins with beta-(thienopyrrolyl)alanines as alternative chromophores and pharmaceutically active amino acids.
Budisa N, Alefelder S, Bae JH, Golbik R, Minks C, Huber R, Moroder L. Budisa N, et al. Protein Sci. 2001 Jul;10(7):1281-92. doi: 10.1110/ps.51601. Protein Sci. 2001. PMID: 11420430 Free PMC article. - Amino acid sequence repertoire of the bacterial proteome and the occurrence of untranslatable sequences.
Navon SP, Kornberg G, Chen J, Schwartzman T, Tsai A, Puglisi EV, Puglisi JD, Adir N. Navon SP, et al. Proc Natl Acad Sci U S A. 2016 Jun 28;113(26):7166-70. doi: 10.1073/pnas.1606518113. Epub 2016 Jun 15. Proc Natl Acad Sci U S A. 2016. PMID: 27307442 Free PMC article. - Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis.
Akashi H, Gojobori T. Akashi H, et al. Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):3695-700. doi: 10.1073/pnas.062526999. Proc Natl Acad Sci U S A. 2002. PMID: 11904428 Free PMC article. - Computational and experimental approaches to chart the Escherichia coli cell-envelope-associated proteome and interactome.
Díaz-Mejía JJ, Babu M, Emili A. Díaz-Mejía JJ, et al. FEMS Microbiol Rev. 2009 Jan;33(1):66-97. doi: 10.1111/j.1574-6976.2008.00141.x. Epub 2008 Nov 27. FEMS Microbiol Rev. 2009. PMID: 19054114 Free PMC article. Review. - Contribution of structural genomics to understanding the biology of Escherichia coli.
Matte A, Sivaraman J, Ekiel I, Gehring K, Jia Z, Cygler M. Matte A, et al. J Bacteriol. 2003 Jul;185(14):3994-4002. doi: 10.1128/JB.185.14.3994-4002.2003. J Bacteriol. 2003. PMID: 12837772 Free PMC article. Review. No abstract available.
Cited by
- Overcoming Challenges in Engineering the Genetic Code.
Lajoie MJ, Söll D, Church GM. Lajoie MJ, et al. J Mol Biol. 2016 Feb 27;428(5 Pt B):1004-21. doi: 10.1016/j.jmb.2015.09.003. Epub 2015 Sep 5. J Mol Biol. 2016. PMID: 26348789 Free PMC article. Review. - All about that Amide Bond: The Sixth Chemical Protein Synthesis (CPS) Meeting.
Weller CE, Chatterjee C. Weller CE, et al. Chembiochem. 2015 Nov;16(17):2531-6. doi: 10.1002/cbic.201500473. Epub 2015 Oct 12. Chembiochem. 2015. PMID: 26457983 Free PMC article. - Plasticity and Constraints of tRNA Aminoacylation Define Directed Evolution of Aminoacyl-tRNA Synthetases.
Crnković A, Vargas-Rodriguez O, Söll D. Crnković A, et al. Int J Mol Sci. 2019 May 9;20(9):2294. doi: 10.3390/ijms20092294. Int J Mol Sci. 2019. PMID: 31075874 Free PMC article. Review. - Multiomics Analysis Provides Insight into the Laboratory Evolution of Escherichia coli toward the Metabolic Usage of Fluorinated Indoles.
Agostini F, Sinn L, Petras D, Schipp CJ, Kubyshkin V, Berger AA, Dorrestein PC, Rappsilber J, Budisa N, Koksch B. Agostini F, et al. ACS Cent Sci. 2021 Jan 27;7(1):81-92. doi: 10.1021/acscentsci.0c00679. Epub 2020 Nov 20. ACS Cent Sci. 2021. PMID: 33532571 Free PMC article. - Xenomicrobiology: a roadmap for genetic code engineering.
Acevedo-Rocha CG, Budisa N. Acevedo-Rocha CG, et al. Microb Biotechnol. 2016 Sep;9(5):666-76. doi: 10.1111/1751-7915.12398. Epub 2016 Aug 4. Microb Biotechnol. 2016. PMID: 27489097 Free PMC article. Review.
References
- Jukes TH, Osawa S. Comp Biochem Physiol B. 1993;106:489–94. - PubMed
- Moura GR, Paredes Ja, Santos MaS. FEBS Lett. 2010;584:334–41. - PubMed
- Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. Science. 1989;244:182–188. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources