Cysteine-specific ubiquitination protects the peroxisomal import receptor Pex5p against proteasomal degradation - PubMed (original) (raw)
Cysteine-specific ubiquitination protects the peroxisomal import receptor Pex5p against proteasomal degradation
Benjamin Schwartzkopff et al. Biosci Rep. 2015.
Abstract
Peroxisomal matrix protein import is mediated by dynamic import receptors, which cycle between the peroxisomal membrane and the cytosol. Proteins with a type 1 peroxisomal targeting signal (PTS1) are bound by the import receptor Pex5p in the cytosol and guided to the peroxisomal membrane. After cargo translocation into the peroxisomal matrix, the receptor is released from the membrane back to the cytosol in an ATP-dependent manner by the AAA-type ATPases Pex1p and Pex6p. These mechanoenzymes recognize ubiquitinated Pex5p-species as substrates for membrane extraction. The PTS1-receptor is either polyubiquitinated via peptide bonds at two certain lysines and results in proteasomal degradation or monoubiquitinated via a thioester-bond at a conserved cysteine, which enables the recycling of Pex5p and further rounds of matrix protein import. To investigate the physiological relevance of the conserved N-terminal cysteine of Pex5p, the known target amino acids for ubiquitination were substituted by site-directed mutagenesis. In contrast with Pex5pC6A, Pex5pC6K turned out to be functional in PTS1 import and utilization of oleic acid, independent of the lysines at position 18 and 24. In contrast with wild-type Pex5p, Pex5pC6K displays an ubiquitination pattern, similar to the polyubiquitination pattern of Pex4p or Pex22p mutant strains. Moreover, Pex5pC6K displays a significantly reduced steady-state level when the deubiquitinating enzyme Ubp15p is missing. Thus, our results indicate that not the cysteine residue but the position of ubiquitination is important for Pex5p function. The presence of the cysteine prevents polyubiquitination and rapid degradation of Pex5p.
Keywords: Pex5p; peroxisome biogenesis; protein import; type 1 peroxisomal targeting signal (PTS1)-receptor; ubiquitination.
© 2015 Authors.
Figures
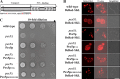
Figure 1. Pex5pC6K can complement a _pex5_Δ strain
(A) Scheme of S. cerevisiae Pex5p. Pex5p contains an N-terminal domain (1–312) with interaction sites required for protein transport and a C-terminal domain (313–612) with six TPR domains (1–6) responsible for the binding of PTS1-proteins. The extreme N-terminus contains the target amino acids for mono- (Cys6) and polyubiquitination (Lys18 and Lys24) highlighted in red. (B) Indicated strains were spotted as a series of 10-fold dilutions on a medium with oleic acid as sole carbon source and incubated for 4 days at 30°C. Mutant _pex5_Δ and _pex5_Δ expressing Pex5pC6A were unable to grow on oleic acid medium. In contrast, the mutant growth phenotype was complemented upon expression of Pex5p, Pex5pK18R/K24R and Pex5pC6K, which display a growth behaviour similar to the wild-type. (C) Oleic acid-induced indicated strains were analysed for the subcellular localization of the transformed PTS1-marker DsRed-SKL by fluorescence microscopy. Mutant _pex5_Δ and _pex5_Δ expressing Pex5pC6A exhibited an overall cellular fluorescence, indicative for a mislocalization of the peroxisomal marker protein as consequence of a peroxisomal import defect. Mutant cells expressing wild-type Pex5p or mutant Pex5pK18R/K24R or Pex5pC6K imported PTS1-proteins properly as indicated by the punctate fluorescence pattern. Scale bar=5 μm
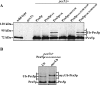
Figure 2. Position-dependent ubiquitination is required for Pex5p function
Mutant _pex5_Δ cells were transformed with plasmids expressing Pex5p or indicated variants. (A) Immunoblot analysis of equal amounts of whole-cell trichloroacetic acid lysates of indicated strains with antibodies against Pex5p. Detection of mitochondrial porin served as loading control. (B) Indicated strains were spotted as a series of 10-fold dilutions on a medium with oleic acid as sole carbon source and incubated for 4 days at 30°C. Growth of mutant _pex5_Δ cells expressing Pex5p or Pex5pK18C was indistinguishable from the wild-type, whereas the mutant _pex5_Δ cells expressing Pex5pC6A/K18C display no growth on this carbon source.
Figure 3. Pex5pC6K is artificially polyubiquitinated independent of K18 and K24
Mutant _pex5_Δ cells were transformed with plasmids expressing Pex5p or indicated variants. (A) Immunoblot analysis of equal amounts of whole-cell trichloroacetic acid lysates of indicated strains with Pex5p-specific antibodies. In contrast with the wild-type, _pex5_Δ expressing Pex5pC6A shows up to three additional αPex5p reactive bands. The third and especially the second band are more pronounced in case of Pex5pC6K and Pex5pC6K/K18R/K24R. (B) The _pex5_Δ strain expressing Pex5pC6K/K18R/K24R was additionally transformed with a plasmid either encoding ubiquitin (Ub) or _myc_-tagged ubiquitin (_myc_Ub). The observed shift to a higher molecular mass upon _myc_Ub expression proved that the additional αPex5p-reactive bands represent ubiquitinated Pex5p.
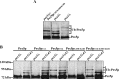
Figure 4. Ubiquitination profile of Pex5pC6K/K18R/K24R is altered by Pex4p
Immunoblot analysis of whole-cell trichloroacetic acid lysates of indicated strains with specific Pex5p antibodies. (A) Mutant _pex5_Δ cells were transformed with plasmids expressing Pex5pC6K/K18R/K24R. The ubiquitination pattern of the Pex5p-variant expressed in mutant _pex5_Δ cells is different from the Pex5p-modification pattern of _pex1_Δ cells, but resembles the pattern for Pex5p of a _pex4_Δ strain, although with shifted intensities. (B) Ubiquitination pattern of indicated Pex5p variants expressed in mutant _pex5_Δ and in double-mutant _pex5_Δ_pex4_Δ cells. The _pex4_Δ-ubiquitination pattern is observed for Pex5p variants when Pex4p is absent, but is more pronounced than in the wild-type. In the Pex5pC6K variants, the second Ub-Pex5p band from below shows a small shift to a lower molecular mass.
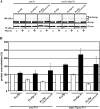
Figure 5. Artificial ubiquitination of Pex5pC6K is linked to higher turnover rate
Mutant _pex5_Δ cells or _ubp15_Δ_pex5_Δ cells were transformed with plasmids expressing Pex5p or indicated variants. (A) Whole-cell lysates of oleic acid-induced strains as indicated were subjected to immunoblot analysis with Pex5p-specific antibodies. Mitochondrial Porin served as control for equal loading. (B) Signal intensities of Pex5p of three independent experiments were estimated by densitometry. Obtained signal intensities were normalized to signal from plasmid-encoded wild-type Pex5p expressed in _pex5_Δ cells. The steady-state concentration of Pex5pC6K and especially Pex5pC6K/K18R/K24R was drastically reduced in the absence of Ubp15p. Error bars=S.E.M.
Figure 6. Rapid proteasomal degradation of Pex5pC6K variants
Mutant _pex5_Δ cells or _ubp15_Δ_pex5_Δ cells were transformed with plasmids expressing Pex5p or indicated variants. Strains were grown on oleic acid containing medium for 16 h. Subsequently cells where shifted to oleic acid containing medium with or without 40 μm MG132 to inhibit the proteasome and grown for additional 4 h. (A) Whole-cell lysates of the strains were prepared and blotted for the presence of Pex5p and mitochondrial porin which served as loading control. (B) Pex5p signal intensities of blots depicted in (A) obtained from samples without (white boxes) and with (black boxes) MG132 treatment was estimated by densitometry. Intensities were normalized to signal from plasmid-encoded wild-type Pex5p expressed in _pex5_Δ cells or ubp15Δ pex5Δ cells respectively, without MG132 treatment. In contrast with untreated cells, Pex5pC6K and Pex5pC6K/K18R/K24R remained stable when the proteasome was inhibited, indicating that the observed reduced steady-state concentrations are due to a rapid proteasomal degradation. Error bars=S.E.M. with n = 3.
Similar articles
- The cytosolic domain of Pex22p stimulates the Pex4p-dependent ubiquitination of the PTS1-receptor.
El Magraoui F, Schrötter A, Brinkmeier R, Kunst L, Mastalski T, Müller T, Marcus K, Meyer HE, Girzalsky W, Erdmann R, Platta HW. El Magraoui F, et al. PLoS One. 2014 Aug 27;9(8):e105894. doi: 10.1371/journal.pone.0105894. eCollection 2014. PLoS One. 2014. PMID: 25162638 Free PMC article. - The deubiquitination of the PTS1-import receptor Pex5p is required for peroxisomal matrix protein import.
El Magraoui F, Brinkmeier R, Mastalski T, Hupperich A, Strehl C, Schwerter D, Girzalsky W, Meyer HE, Warscheid B, Erdmann R, Platta HW. El Magraoui F, et al. Biochim Biophys Acta Mol Cell Res. 2019 Feb;1866(2):199-213. doi: 10.1016/j.bbamcr.2018.11.002. Epub 2018 Nov 6. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 30408545 - Ubiquitination of the peroxisomal import receptor Pex5p.
Platta HW, Girzalsky W, Erdmann R. Platta HW, et al. Biochem J. 2004 Nov 15;384(Pt 1):37-45. doi: 10.1042/BJ20040572. Biochem J. 2004. PMID: 15283676 Free PMC article. - New insights into dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p in shuttling of PTS1-receptor Pex5p during peroxisome biogenesis.
Fujiki Y, Nashiro C, Miyata N, Tamura S, Okumoto K. Fujiki Y, et al. Biochim Biophys Acta. 2012 Jan;1823(1):145-9. doi: 10.1016/j.bbamcr.2011.10.012. Epub 2011 Nov 4. Biochim Biophys Acta. 2012. PMID: 22079764 Review. - The AAA-type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in Saccharomyces cerevisiae.
Grimm I, Saffian D, Platta HW, Erdmann R. Grimm I, et al. Biochim Biophys Acta. 2012 Jan;1823(1):150-8. doi: 10.1016/j.bbamcr.2011.09.005. Epub 2011 Sep 22. Biochim Biophys Acta. 2012. PMID: 21963882 Review.
Cited by
- Ceramide regulates interaction of Hsd17b4 with Pex5 and function of peroxisomes.
Zhu Z, Chen J, Wang G, Elsherbini A, Zhong L, Jiang X, Qin H, Tripathi P, Zhi W, Spassieva SD, Morris AJ, Bieberich E. Zhu Z, et al. Biochim Biophys Acta Mol Cell Biol Lipids. 2019 Oct;1864(10):1514-1524. doi: 10.1016/j.bbalip.2019.05.017. Epub 2019 Jun 5. Biochim Biophys Acta Mol Cell Biol Lipids. 2019. PMID: 31176039 Free PMC article. - The peroxisome protein translocation machinery is developmentally regulated in the fungus Podospora anserina.
Aguirre-López B, Suaste-Olmos F, Peraza-Reyes L. Aguirre-López B, et al. Microbiol Spectr. 2024 Jan 11;12(1):e0213923. doi: 10.1128/spectrum.02139-23. Epub 2023 Dec 13. Microbiol Spectr. 2024. PMID: 38088545 Free PMC article. - Post-translational modifications of proteins associated with yeast peroxisome membrane: An essential mode of regulatory mechanism.
Infant T, Deb R, Ghose S, Nagotu S. Infant T, et al. Genes Cells. 2021 Nov;26(11):843-860. doi: 10.1111/gtc.12892. Epub 2021 Sep 2. Genes Cells. 2021. PMID: 34472666 Free PMC article. Review. - Peroxisome protein import: a complex journey.
Baker A, Lanyon-Hogg T, Warriner SL. Baker A, et al. Biochem Soc Trans. 2016 Jun 15;44(3):783-9. doi: 10.1042/BST20160036. Biochem Soc Trans. 2016. PMID: 27284042 Free PMC article. Review. - Current Advances in Protein Import into Peroxisomes.
Walter T, Erdmann R. Walter T, et al. Protein J. 2019 Jun;38(3):351-362. doi: 10.1007/s10930-019-09835-6. Protein J. 2019. PMID: 31054036 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases