Self-guided Langevin dynamics via generalized Langevin equation - PubMed (original) (raw)
. 2016 Mar 5;37(6):595-601.
doi: 10.1002/jcc.24015. Epub 2015 Jul 16.
Affiliations
- PMID: 26183423
- PMCID: PMC4715807
- DOI: 10.1002/jcc.24015
Self-guided Langevin dynamics via generalized Langevin equation
Xiongwu Wu et al. J Comput Chem. 2016.
Abstract
Self-guided Langevin dynamics (SGLD) is a molecular simulation method that enhances conformational search and sampling via acceleration of the low frequency motions of the system. This acceleration is produced via introduction of a guiding force which breaks down the detailed-balance property of the dynamics, implying that some reweighting is necessary to perform equilibrium sampling. Here, we eliminate the need of reweighing and show that the NVT and NPT ensembles are sampled exactly by a new version of self-guided motion involving a generalized Langevin equation (GLE) in which the random force is modified so as to restore detailed-balance. Through the examples of alanine dipeptide and argon liquid, we show that this SGLD-GLE method has enhanced conformational sampling capabilities compared with regular Langevin dynamics (LD) while being of comparable computational complexity. In particular, SGLD-GLE is fully size extensive and can be used in arbitrarily large systems, making it an appealing alternative to LD. © 2015 Wiley Periodicals, Inc.
Keywords: canonical ensemble; conformational sampling; generalized Langevin equation; molecular simulation; self-guided Langevin dynamics.
© 2015 Wiley Periodicals, Inc.
Figures
Fig.1
An alanine dipeptide and its motions in LD and SGLD simulations. Motions are marked by colored arrays with blue, yellow, and red to represent cold, normal, and hot temperatures. _T_lf and _T_hf represent low frequency temperature and high frequency temperature, respectively.
Fig.2
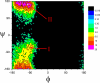
the ϕ-φ distributions of an alanine dipeptide obtained from a 20 ns LD simulation at _T_=300 K, _ξ_=10/ps. Two popular regions, I around (−90°,−70°) and II around (−90°, 160°), are identified.
Fig.3
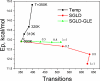
Average potential energies of the alanine dipeptide vs transitions between the the two ϕ–φ regions in high temperature LD, SGLD, and SGLD-GLE simulations. All the SGLD and SGLDGLE simulations were performed at _T_=300K with _ξ_=10/ps.
Fig.4
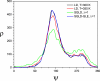
The φ angle distributions of the alanine dipeptide in the LD, SGLD, and SGLD-GLE simulations. All simulations were performed at _T_=300K, _ξ_=10/ps except the high temperature LD performed at _T_=350 K.
Fig.5
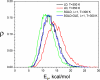
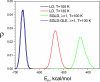
The potential energy distribution of the alanine dipeptide in LD at T=300K and T=350K, SGLD, and SGLD-GLE simulations at T=300K, _ξ_=10/ps, and _λ_=1.
Fig.6
The potential energy distributions of the liquid argon from LD, SGLD, and SGLD-GLE simulations. All simulations were performed at _T_=100 K, _ξ_=10/ps, except that the high temperature LD simulation that was performed at _T_=150 K. The guiding factor was λ=1 for both the SGLD and SGLD-GLE simulations.
Fig.7
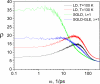
The average potential energies and volumes of argon liquid are plot against the diffusion constants in high temperature LD, SGLD, and SGLD-GLE simulations. All simulations were performed at ξ=10/ps, T=100 K and P=1 atm except labeled otherwise. The high-temperature LD simulations are labeled with temperatures. The SGLD and SGLD-GLE simulations were labelled with the guiding factors.
Fig.8
The spectrum of the argon liquid from LD, SGLD, and SGLD-GLE simulations. All simulations were performed at T=100 K, P=1 atm, and ξ=10/ps, except that the high temperature LD was performed at _T_=150 K. The guiding factor was λ=1 for the SGLD and SGLD-GLE simulations.
Similar articles
- Toward canonical ensemble distribution from self-guided Langevin dynamics simulation.
Wu X, Brooks BR. Wu X, et al. J Chem Phys. 2011 Apr 7;134(13):134108. doi: 10.1063/1.3574397. J Chem Phys. 2011. PMID: 21476744 Free PMC article. - Force-momentum-based self-guided Langevin dynamics: a rapid sampling method that approaches the canonical ensemble.
Wu X, Brooks BR. Wu X, et al. J Chem Phys. 2011 Nov 28;135(20):204101. doi: 10.1063/1.3662489. J Chem Phys. 2011. PMID: 22128922 Free PMC article. - The multi-dimensional generalized Langevin equation for conformational motion of proteins.
Lee HS, Ahn SH, Darve EF. Lee HS, et al. J Chem Phys. 2019 May 7;150(17):174113. doi: 10.1063/1.5055573. J Chem Phys. 2019. PMID: 31067888 - Structural Fluctuation, Relaxation, and Folding of Protein: An Approach Based on the Combined Generalized Langevin and RISM/3D-RISM Theories.
Hirata F. Hirata F. Molecules. 2023 Oct 30;28(21):7351. doi: 10.3390/molecules28217351. Molecules. 2023. PMID: 37959769 Free PMC article. Review. - Enhanced sampling algorithms.
Mitsutake A, Mori Y, Okamoto Y. Mitsutake A, et al. Methods Mol Biol. 2013;924:153-95. doi: 10.1007/978-1-62703-017-5_7. Methods Mol Biol. 2013. PMID: 23034749 Review.
Cited by
- DFMD: Fast and Effective DelPhiForce Steered Molecular Dynamics Approach to Model Ligand Approach Toward a Receptor: Application to Spermine Synthase Enzyme.
Peng Y, Yang Y, Li L, Jia Z, Cao W, Alexov E. Peng Y, et al. Front Mol Biosci. 2019 Sep 4;6:74. doi: 10.3389/fmolb.2019.00074. eCollection 2019. Front Mol Biosci. 2019. PMID: 31552265 Free PMC article. - Deep Boosted Molecular Dynamics (DBMD): Accelerating molecular simulations with Gaussian boost potentials generated using probabilistic Bayesian deep neural network.
Do HN, Miao Y. Do HN, et al. bioRxiv [Preprint]. 2023 Apr 5:2023.03.25.534210. doi: 10.1101/2023.03.25.534210. bioRxiv. 2023. PMID: 37034713 Free PMC article. Updated. Preprint. - Molecular dynamics simulations of lipid nanodiscs.
Pourmousa M, Pastor RW. Pourmousa M, et al. Biochim Biophys Acta Biomembr. 2018 Oct;1860(10):2094-2107. doi: 10.1016/j.bbamem.2018.04.015. Epub 2018 May 3. Biochim Biophys Acta Biomembr. 2018. PMID: 29729280 Free PMC article. - Per- and Polyfluoroalkyl (PFAS) Disruption of Thyroid Hormone Synthesis.
Bali SK, Martin R, Almeida NMS, Saunders C, Wilson AK. Bali SK, et al. ACS Omega. 2024 Sep 10;9(38):39554-39563. doi: 10.1021/acsomega.4c03578. eCollection 2024 Sep 24. ACS Omega. 2024. PMID: 39346893 Free PMC article. - Adaptive Synthesis of Functional Amphiphilic Dendrons as a Novel Approach to Artificial Supramolecular Objects.
Edr A, Wrobel D, Krupková A, Šťastná LČ, Cuřínová P, Novák A, Malý J, Kalasová J, Malý J, Malý M, Strašák T. Edr A, et al. Int J Mol Sci. 2022 Feb 14;23(4):2114. doi: 10.3390/ijms23042114. Int J Mol Sci. 2022. PMID: 35216229 Free PMC article.
References
- Wu X, Brooks BR. Chemical Physics Letters. 2003;381:512–518.
- Damjanovic A, Miller BT, Wenaus TJ, Maksimovic P, Bertrand Garcia-Moreno E, Brooks BR. Journal of Chemical Information and Modeling. 2008;48:2021–2029. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials