QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments - PubMed (original) (raw)
QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments
Stephen W Hartley et al. BMC Bioinformatics. 2015.
Abstract
Background: High-throughput next-generation RNA sequencing has matured into a viable and powerful method for detecting variations in transcript expression and regulation. Proactive quality control is of critical importance as unanticipated biases, artifacts, or errors can potentially drive false associations and lead to flawed results.
Results: We have developed the Quality of RNA-Seq Toolset, or QoRTs, a comprehensive, multifunction toolset that assists in quality control and data processing of high-throughput RNA sequencing data.
Conclusions: QoRTs generates an unmatched variety of quality control metrics, and can provide cross-comparisons of replicates contrasted by batch, biological sample, or experimental condition, revealing any outliers and/or systematic issues that could drive false associations or otherwise compromise downstream analyses. In addition, QoRTs simultaneously replaces the functionality of numerous other data-processing tools, and can quickly and efficiently generate quality control metrics, coverage counts (for genes, exons, and known/novel splice-junctions), and browser tracks. These functions can all be carried out as part of a single unified data-processing/quality control run, greatly reducing both the complexity and the total runtime of the analysis pipeline. The software, source code, and documentation are available online at http://hartleys.github.io/QoRTs.
Figures
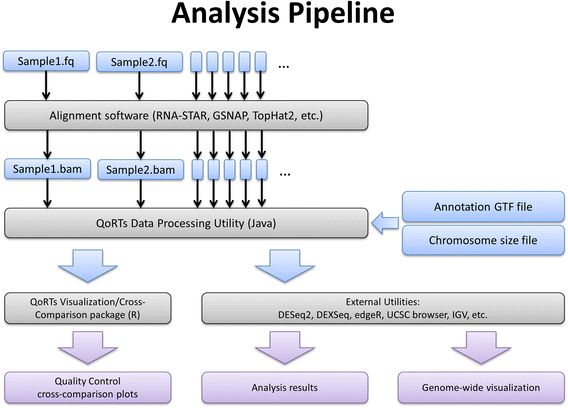
Fig. 1
An example analysis pipeline with QoRTs. This flowchart illustrates the recommended analysis pipeline for conventional RNA-Seq analysis using QoRTs. Input and intermediary files are shown in blue, output files and results are shown in purple
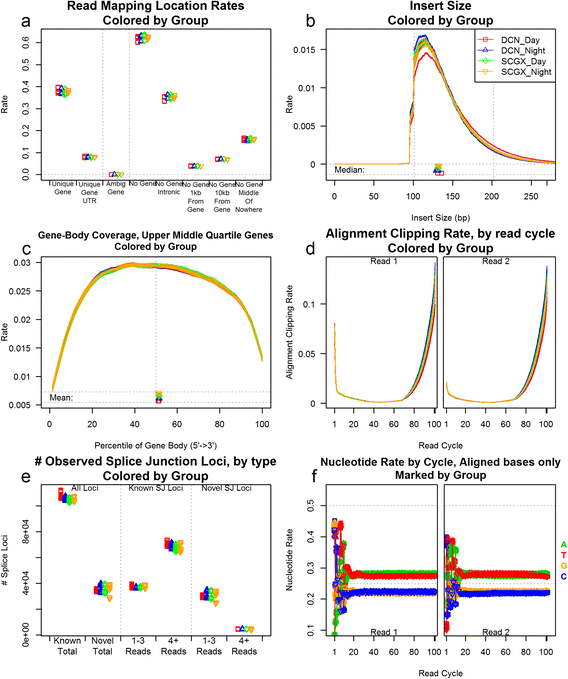
Fig. 2
A small selection of the QC plots offered by QoRTs. This series includes 12 samples, each consisting of 6 technical replicates (for a total of 72 bam files), with 4 different biological conditions (3 samples per condition). In all nine plots, replicates are colored and differentiated by biological group. In the line plots (c,d,e, and f) the samples are simply colored by biological group. In other plots (a and g), replicates are differentiated by character, color, and horizontal offset. This differentiation allows easy identification of both outliers and systematic biases or errors associated with the biological condition. Such systematic errors are of particular importance as they could potentially drive false associations. A full description of each plot and its interpretation can be found in the supplementary materials
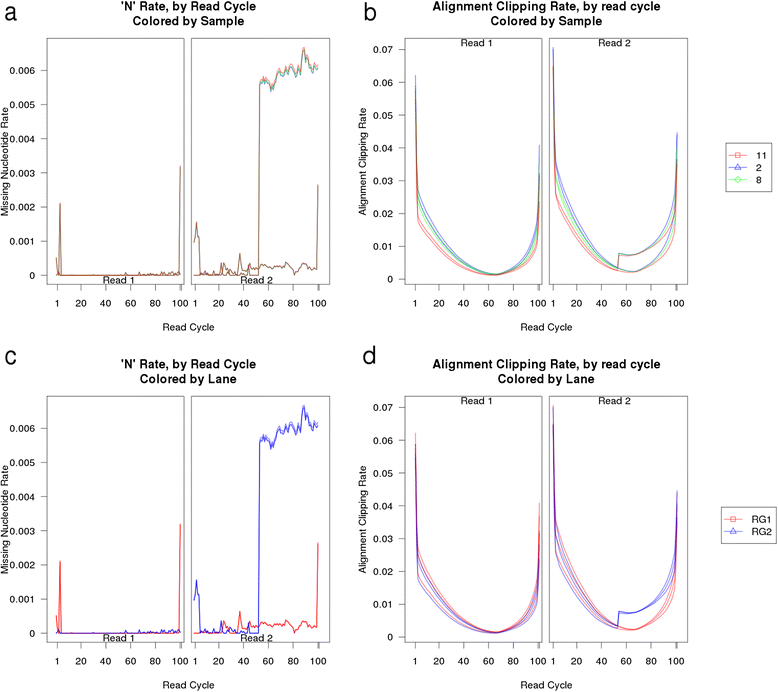
Fig. 3
Example issue detected via QoRTs. A subset of the output plots from a dataset in which a rare hardware-level fault produced an actionable QC issue that can be easily identified via QoRTs. In (a) and (b) the replicates are colored by biological sample; in (c) and (d) replicates are colored by sequencer lane. See the QoRTs vignette for more information (Additional file 1)
Similar articles
- RNA-QC-chain: comprehensive and fast quality control for RNA-Seq data.
Zhou Q, Su X, Jing G, Chen S, Ning K. Zhou Q, et al. BMC Genomics. 2018 Feb 14;19(1):144. doi: 10.1186/s12864-018-4503-6. BMC Genomics. 2018. PMID: 29444661 Free PMC article. - Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM).
Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, Stoeckert CJ, Hogenesch JB, Pierce EA. Grant GR, et al. Bioinformatics. 2011 Sep 15;27(18):2518-28. doi: 10.1093/bioinformatics/btr427. Epub 2011 Jul 19. Bioinformatics. 2011. PMID: 21775302 Free PMC article. - iSeqQC: a tool for expression-based quality control in RNA sequencing.
Kumar G, Ertel A, Feldman G, Kupper J, Fortina P. Kumar G, et al. BMC Bioinformatics. 2020 Feb 13;21(1):56. doi: 10.1186/s12859-020-3399-8. BMC Bioinformatics. 2020. PMID: 32054449 Free PMC article. - Snaptron: querying splicing patterns across tens of thousands of RNA-seq samples.
Wilks C, Gaddipati P, Nellore A, Langmead B. Wilks C, et al. Bioinformatics. 2018 Jan 1;34(1):114-116. doi: 10.1093/bioinformatics/btx547. Bioinformatics. 2018. PMID: 28968689 Free PMC article. - Differential Expression Analysis of RNA-seq Reads: Overview, Taxonomy, and Tools.
Chowdhury HA, Bhattacharyya DK, Kalita JK. Chowdhury HA, et al. IEEE/ACM Trans Comput Biol Bioinform. 2020 Mar-Apr;17(2):566-586. doi: 10.1109/TCBB.2018.2873010. Epub 2018 Oct 1. IEEE/ACM Trans Comput Biol Bioinform. 2020. PMID: 30281477 Review.
Cited by
- Local and systemic transcriptome and spliceome reprogramming induced by the root-knot nematode Meloidogyne incognita in tomato.
Ozdemir S, Piya S, Lopes-Caitar VS, Coffey N, Rice JH, Hewezi T. Ozdemir S, et al. Hortic Res. 2024 Jul 26;11(9):uhae206. doi: 10.1093/hr/uhae206. eCollection 2024 Sep. Hortic Res. 2024. PMID: 39286358 Free PMC article. - Transcriptomic analysis of the spatiotemporal axis of oogenesis and fertilization in C. elegans.
Su Y, Shea J, Destephanis D, Su Z. Su Y, et al. Front Cell Dev Biol. 2024 Aug 19;12:1436975. doi: 10.3389/fcell.2024.1436975. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39224437 Free PMC article. - Scalable Hypothalamic Arcuate Neuron Differentiation from Human Pluripotent Stem Cells Suitable for Modeling Metabolic and Reproductive Disorders.
Jovanovic VM, Narisu N, Bonnycastle LL, Tharakan R, Mesch KT, Glover HJ, Yan T, Sinha N, Sen C, Castellano D, Yang S, Blivis D, Ryu S, Bennett DF, Rosales-Soto G, Inman J, Ormanoglu P, LeClair CA, Xia M, Schneider M, Hernandez-Ochoa EO, Erdos MR, Simeonov A, Chen S, Collins FS, Doege CA, Tristan CA. Jovanovic VM, et al. bioRxiv [Preprint]. 2024 Sep 20:2024.06.27.601062. doi: 10.1101/2024.06.27.601062. bioRxiv. 2024. PMID: 39005353 Free PMC article. Preprint. - Transcriptomic Analysis of the Spatiotemporal Axis of Oogenesis and Fertilization in C. elegans.
Su Y, Shea J, DeStephanis D, Su Z. Su Y, et al. bioRxiv [Preprint]. 2024 Jun 4:2024.06.03.597235. doi: 10.1101/2024.06.03.597235. bioRxiv. 2024. PMID: 38895354 Free PMC article. Updated. Preprint. - The fungicide pyraclostrobin affects gene expression by altering the DNA methylation pattern in Magnaporthe oryzae.
Fang S, Wang H, Qiu K, Pang Y, Li C, Liang X. Fang S, et al. Front Plant Sci. 2024 Apr 30;15:1391900. doi: 10.3389/fpls.2024.1391900. eCollection 2024. Front Plant Sci. 2024. PMID: 38745924 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources