Strain-resolved microbial community proteomics reveals simultaneous aerobic and anaerobic function during gastrointestinal tract colonization of a preterm infant - PubMed (original) (raw)
Strain-resolved microbial community proteomics reveals simultaneous aerobic and anaerobic function during gastrointestinal tract colonization of a preterm infant
Brandon Brooks et al. Front Microbiol. 2015.
Abstract
While there has been growing interest in the gut microbiome in recent years, it remains unclear whether closely related species and strains have similar or distinct functional roles and if organisms capable of both aerobic and anaerobic growth do so simultaneously. To investigate these questions, we implemented a high-throughput mass spectrometry-based proteomics approach to identify proteins in fecal samples collected on days of life 13-21 from an infant born at 28 weeks gestation. No prior studies have coupled strain-resolved community metagenomics to proteomics for such a purpose. Sequences were manually curated to resolve the genomes of two strains of Citrobacter that were present during the later stage of colonization. Proteome extracts from fecal samples were processed via a nano-2D-LC-MS/MS and peptides were identified based on information predicted from the genome sequences for the dominant organisms, Serratia and the two Citrobacter strains. These organisms are facultative anaerobes, and proteomic information indicates the utilization of both aerobic and anaerobic metabolisms throughout the time series. This may indicate growth in distinct niches within the gastrointestinal tract. We uncovered differences in the physiology of coexisting Citrobacter strains, including differences in motility and chemotaxis functions. Additionally, for both Citrobacter strains we resolved a community-essential role in vitamin metabolism and a predominant role in propionate production. Finally, in this case study we detected differences between genome abundance and activity levels for the dominant populations. This underlines the value in layering proteomic information over genetic potential.
Keywords: colonization; infant gut; metaproteomics; microbial ecology; microbiome; physiology.
Figures
Figure 1
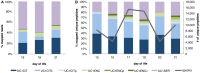
Microbial community composition observed via read and peptide mapping. Relative proportion of reads (A) and unique peptides (spectral counts normalized by protein length and number of proteins per genome for each sample) (B) mapped to a database of metagenomes derived from dominant gut colonizers in a preterm infant. Abbreviations for each organism begin with UC1 (University of Chicago, study code 1), followed by the organism name: CIT, Citrobacter; CITii, Citrobacter minor strain; CITp, Citrobacter plasmid; ENC, Enterococcus; ENCp, Enterococcus plasmid; ENCv, Enterococcus phage; SER, Serratia.
Figure 2
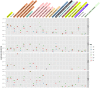
Metabolic potential of microbes colonizing a preterm infant gut. ggKbase lists illustrate the broad metabolic potential of microbes colonizing a preterm infant in the first month of life. Numbers in each cell are the number of features per organism per list. Colors per cell are generated in a heat map fashion, with darker colors representing more features per list relative to other organisms.
Figure 3
Expression over potential (genomic content) ratio of infant gut microbes. A non-redundant count of the number of features identified via proteomics in a metabolic ggKbase list was divided by the number of features in that list and plotted for each organism across time.
Figure 4
Comparison of proteomic profiles of two closely related Citrobacter strains. Unique peptide counts, normalized by the length of the protein, were mapped to UC1CITs' contigs (aligned on the x-axis). Panels are separated by day of life. For a description of ggKbase lists, see the Materials and Methods and Supplemental File 1. Triangles represent genes unique to the respective strain. Annotations marked with arrows indicate features of interest.
Similar articles
- Genome-resolved metaproteomic characterization of preterm infant gut microbiota development reveals species-specific metabolic shifts and variabilities during early life.
Xiong W, Brown CT, Morowitz MJ, Banfield JF, Hettich RL. Xiong W, et al. Microbiome. 2017 Jul 10;5(1):72. doi: 10.1186/s40168-017-0290-6. Microbiome. 2017. PMID: 28693612 Free PMC article. - Hospitalized Premature Infants Are Colonized by Related Bacterial Strains with Distinct Proteomic Profiles.
Brown CT, Xiong W, Olm MR, Thomas BC, Baker R, Firek B, Morowitz MJ, Hettich RL, Banfield JF. Brown CT, et al. mBio. 2018 Apr 10;9(2):e00441-18. doi: 10.1128/mBio.00441-18. mBio. 2018. PMID: 29636439 Free PMC article. - Strain-resolved community genomic analysis of gut microbial colonization in a premature infant.
Morowitz MJ, Denef VJ, Costello EK, Thomas BC, Poroyko V, Relman DA, Banfield JF. Morowitz MJ, et al. Proc Natl Acad Sci U S A. 2011 Jan 18;108(3):1128-33. doi: 10.1073/pnas.1010992108. Epub 2010 Dec 29. Proc Natl Acad Sci U S A. 2011. PMID: 21191099 Free PMC article. - Metaproteomics as a Complementary Approach to Gut Microbiota in Health and Disease.
Petriz BA, Franco OL. Petriz BA, et al. Front Chem. 2017 Jan 26;5:4. doi: 10.3389/fchem.2017.00004. eCollection 2017. Front Chem. 2017. PMID: 28184370 Free PMC article. Review. - Rumen metaproteomics: Closer to linking rumen microbial function to animal productivity traits.
Andersen TO, Kunath BJ, Hagen LH, Arntzen MØ, Pope PB. Andersen TO, et al. Methods. 2021 Feb;186:42-51. doi: 10.1016/j.ymeth.2020.07.011. Epub 2020 Aug 3. Methods. 2021. PMID: 32758682 Review.
Cited by
- The astounding exhaustiveness and speed of the Astral mass analyzer for highly complex samples is a quantum leap in the functional analysis of microbiomes.
Dumas T, Martinez Pinna R, Lozano C, Radau S, Pible O, Grenga L, Armengaud J. Dumas T, et al. Microbiome. 2024 Mar 7;12(1):46. doi: 10.1186/s40168-024-01766-4. Microbiome. 2024. PMID: 38454512 Free PMC article. - Considerations for constructing a protein sequence database for metaproteomics.
Blakeley-Ruiz JA, Kleiner M. Blakeley-Ruiz JA, et al. Comput Struct Biotechnol J. 2022 Jan 21;20:937-952. doi: 10.1016/j.csbj.2022.01.018. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35242286 Free PMC article. Review. - Similar Strains of Coagulase-Negative Staphylococci Found in the Gastrointestinal Tract and Bloodstream of Bacteremic Neonates.
Adeghate JO, Juhász E, Iván MÁ, Pongrácz J, Kristóf K. Adeghate JO, et al. Can J Infect Dis Med Microbiol. 2020 Jul 21;2020:3509676. doi: 10.1155/2020/3509676. eCollection 2020. Can J Infect Dis Med Microbiol. 2020. PMID: 32774563 Free PMC article. - Estimating relative biomasses of organisms in microbiota using "phylopeptidomics".
Pible O, Allain F, Jouffret V, Culotta K, Miotello G, Armengaud J. Pible O, et al. Microbiome. 2020 Mar 6;8(1):30. doi: 10.1186/s40168-020-00797-x. Microbiome. 2020. PMID: 32143687 Free PMC article. - Combined analysis of microbial metagenomic and metatranscriptomic sequencing data to assess in situ physiological conditions in the premature infant gut.
Sher Y, Olm MR, Raveh-Sadka T, Brown CT, Sher R, Firek B, Baker R, Morowitz MJ, Banfield JF. Sher Y, et al. PLoS One. 2020 Mar 4;15(3):e0229537. doi: 10.1371/journal.pone.0229537. eCollection 2020. PLoS One. 2020. PMID: 32130257 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources