Randomized Trial of Rapid Multiplex Polymerase Chain Reaction-Based Blood Culture Identification and Susceptibility Testing - PubMed (original) (raw)
Randomized Controlled Trial
. 2015 Oct 1;61(7):1071-80.
doi: 10.1093/cid/civ447. Epub 2015 Jul 20.
Affiliations
- PMID: 26197846
- PMCID: PMC4560903
- DOI: 10.1093/cid/civ447
Randomized Controlled Trial
Randomized Trial of Rapid Multiplex Polymerase Chain Reaction-Based Blood Culture Identification and Susceptibility Testing
Ritu Banerjee et al. Clin Infect Dis. 2015.
Abstract
Background: The value of rapid, panel-based molecular diagnostics for positive blood culture bottles (BCBs) has not been rigorously assessed. We performed a prospective randomized controlled trial evaluating outcomes associated with rapid multiplex PCR (rmPCR) detection of bacteria, fungi, and resistance genes directly from positive BCBs.
Methods: A total of 617 patients with positive BCBs underwent stratified randomization into 3 arms: standard BCB processing (control, n = 207), rmPCR reported with templated comments (rmPCR, n = 198), or rmPCR reported with templated comments and real-time audit and feedback of antimicrobial orders by an antimicrobial stewardship team (rmPCR/AS, n = 212). The primary outcome was antimicrobial therapy duration. Secondary outcomes were time to antimicrobial de-escalation or escalation, length of stay (LOS), mortality, and cost.
Results: Time from BCB Gram stain to microorganism identification was shorter in the intervention group (1.3 hours) vs control (22.3 hours) (P < .001). Compared to the control group, both intervention groups had decreased broad-spectrum piperacillin-tazobactam (control 56 hours, rmPCR 44 hours, rmPCR/AS 45 hours; P = .01) and increased narrow-spectrum β-lactam (control 42 hours, rmPCR 71 hours, rmPCR/AS 85 hours; P = .04) use, and less treatment of contaminants (control 25%, rmPCR 11%, rmPCR/AS 8%; P = .015). Time from Gram stain to appropriate antimicrobial de-escalation or escalation was shortest in the rmPCR/AS group (de-escalation: rmPCR/AS 21 hours, control 34 hours, rmPCR 38 hours, P < .001; escalation: rmPCR/AS 5 hours, control 24 hours, rmPCR 6 hours, P = .04). Groups did not differ in mortality, LOS, or cost.
Conclusions: rmPCR reported with templated comments reduced treatment of contaminants and use of broad-spectrum antimicrobials. Addition of antimicrobial stewardship enhanced antimicrobial de-escalation.
Clinical trials registration: NCT01898208.
Keywords: PCR; antimicrobial stewardship; blood culture; diagnostic.
© The Author 2015. Published by Oxford University Press on behalf of the Infectious Diseases Society of America. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
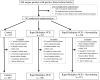
Figure 1.
Participant enrollment. System/technical errors included randomization software downtime, typographical errors causing an ineligible subject to be erroneously randomized, and failure to enter an eligible subject into the randomization program. Abbreviation: PCR, polymerase chain reaction.
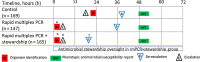
Figure 2.
Comparison of time to organism identification, availability of phenotypic antimicrobial susceptibility results, and first appropriate modification of antimicrobial therapy for the subset of study subjects with organisms represented on the rapid multiplex polymerase chain reaction (rmPCR) panel (n = 481). Time 0 is when the positive Gram stain result was reported. Median time in hours (interquartile range [IQR]) to organism identification: control 22.3 (17–28), both rmPCR and rmPCR + stewardship 1.3 (0.9–1.6); de-escalation: control 39 (19–56), rmPCR 36 (22–61), rmPCR + stewardship 20 (6–36); escalation: control 18 (2–63), rmPCR 4 (1.5–24), rmPCR + stewardship 4 (1.8–9). *P < .05 vs control; †P < .05 vs control and rmPCR groups.
Comment in
- Editorial Commentary: Rapid Blood Culture Identification: The Value of a Randomized Trial.
Caliendo AM. Caliendo AM. Clin Infect Dis. 2015 Oct 1;61(7):1081-3. doi: 10.1093/cid/civ450. Epub 2015 Jul 20. Clin Infect Dis. 2015. PMID: 26197845 No abstract available. - Reply to Idelevich and Beck.
Banerjee R, Teng CB, Cunningham SA, Ihde S, Steckelberg JP, Moriarty JP, Shah ND, Mandrekar J, Patel R. Banerjee R, et al. Clin Infect Dis. 2016 Jan 15;62(2):269-70. doi: 10.1093/cid/civ826. Epub 2015 Sep 22. Clin Infect Dis. 2016. PMID: 26394668 No abstract available. - Identification and Susceptibility Testing From Shortly Incubated Cultures Accelerate Blood Culture Diagnostics at No Cost.
Idelevich EA, Becker K. Idelevich EA, et al. Clin Infect Dis. 2016 Jan 15;62(2):268-9. doi: 10.1093/cid/civ824. Epub 2015 Sep 22. Clin Infect Dis. 2016. PMID: 26394670 No abstract available.
Similar articles
- Impact of a rapid multiplex polymerase chain reaction blood culture identification technology on outcomes in patients with vancomycin-resistant Enterococcal bacteremia.
MacVane SH, Hurst JM, Boger MS, Gnann JW Jr. MacVane SH, et al. Infect Dis (Lond). 2016 Oct;48(10):732-7. doi: 10.1080/23744235.2016.1185533. Epub 2016 May 19. Infect Dis (Lond). 2016. PMID: 27196015 - Benefits of Adding a Rapid PCR-Based Blood Culture Identification Panel to an Established Antimicrobial Stewardship Program.
MacVane SH, Nolte FS. MacVane SH, et al. J Clin Microbiol. 2016 Oct;54(10):2455-63. doi: 10.1128/JCM.00996-16. Epub 2016 Aug 3. J Clin Microbiol. 2016. PMID: 27487951 Free PMC article. - Rapid multiplex PCR panel for pneumonia in hospitalised patients with suspected pneumonia in the USA: a single-centre, open-label, pragmatic, randomised controlled trial.
Virk A, Strasburg AP, Kies KD, Donadio AD, Mandrekar J, Harmsen WS, Stevens RW, Estes LL, Tande AJ, Challener DW, Osmon DR, Fida M, Vergidis P, Suh GA, Wilson JW, Rajapakse NS, Borah BJ, Dholakia R, Reed KA, Hines LM, Schuetz AN, Patel R. Virk A, et al. Lancet Microbe. 2024 Dec;5(12):100928. doi: 10.1016/S2666-5247(24)00170-8. Epub 2024 Oct 17. Lancet Microbe. 2024. PMID: 39426396 Clinical Trial.
Cited by
- Updating Molecular Diagnostics for Detecting Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus Isolates in Blood Culture Bottles.
Tenover FC, Tickler IA, Le VM, Dewell S, Mendes RE, Goering RV. Tenover FC, et al. J Clin Microbiol. 2019 Oct 23;57(11):e01195-19. doi: 10.1128/JCM.01195-19. Print 2019 Nov. J Clin Microbiol. 2019. PMID: 31484703 Free PMC article. - Molecular diagnostics for genotypic detection of antibiotic resistance: current landscape and future directions.
Banerjee R, Patel R. Banerjee R, et al. JAC Antimicrob Resist. 2023 Feb 17;5(1):dlad018. doi: 10.1093/jacamr/dlad018. eCollection 2023 Feb. JAC Antimicrob Resist. 2023. PMID: 36816746 Free PMC article. Review. - Validation of an Antimicrobial Stewardship-Driven Verigene Blood-Culture Gram-Negative Treatment Algorithm to Improve Appropriateness of Antibiotics.
Claeys KC, Schlaffer KE, Heil EL, Leekha S, Johnson JK. Claeys KC, et al. Open Forum Infect Dis. 2018 Sep 15;5(10):ofy233. doi: 10.1093/ofid/ofy233. eCollection 2018 Oct. Open Forum Infect Dis. 2018. PMID: 30568975 Free PMC article. - Susceptibility Provision Enhances Effective De-escalation (SPEED): utilizing rapid phenotypic susceptibility testing in Gram-negative bloodstream infections and its potential clinical impact.
Schneider JG, Wood JB, Schmitt BH, Emery CL, Davis TE, Smith NW, Blevins S, Hiles J, Desai A, Wrin J, Bocian B, Manaloor JJ. Schneider JG, et al. J Antimicrob Chemother. 2019 Jan 1;74(Suppl 1):i16-i23. doi: 10.1093/jac/dky531. J Antimicrob Chemother. 2019. PMID: 30690542 Free PMC article. - Outpatient Antibiotic and Antiviral Utilization Patterns in Patients Tested for Respiratory Pathogens in the United States: A Real-World Database Study.
Tse J, Near AM, Cheng M, Karichu J, Lee B, Chang SN. Tse J, et al. Antibiotics (Basel). 2022 Aug 4;11(8):1058. doi: 10.3390/antibiotics11081058. Antibiotics (Basel). 2022. PMID: 36009927 Free PMC article.
References
- Ibrahim E, Sherman G, Ward S, Fraser V, Kollef M. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 2000; 118:46–55. - PubMed
- Garnacho-Montero J, Gutierrez-Pizarraya A, Escoresca-Ortega A et al. . De-escalation of empirical therapy is associated with lower mortality in patients with severe sepsis and septic shock. Intensive Care Med 2014; 40:32–40. - PubMed
- Dellit T, Owens R, McGowan J et al. . Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an an institutional program for enhancing antimicrobial stewardship. Clin Infect Dis 2007; 44:159–77. - PubMed
- Perez K, Olsen R, Musick W et al. . Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch Pathol Lab Med 2012; 137:1247–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials