MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures - PubMed (original) (raw)
Santhilal Subhash 1, Roshan Vaid 1, Stefan Enroth 2, Sireesha Uday 1, Björn Reinius 1, Sanhita Mitra 1, Arif Mohammed 1, Alva Rani James 1, Emily Hoberg 3, Aristidis Moustakas 4, Ulf Gyllensten 2, Steven J M Jones 5, Claes M Gustafsson 3, Andrew H Sims 6, Fredrik Westerlund 7, Eduardo Gorab 8, Chandrasekhar Kanduri 1
Affiliations
- PMID: 26205790
- PMCID: PMC4525211
- DOI: 10.1038/ncomms8743
MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures
Tanmoy Mondal et al. Nat Commun. 2015.
Erratum in
- Author Correction: MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures.
Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJM, Gustafsson CM, Sims AH, Westerlund F, Gorab E, Kanduri C. Mondal T, et al. Nat Commun. 2019 Nov 21;10(1):5290. doi: 10.1038/s41467-019-13200-7. Nat Commun. 2019. PMID: 31754097 Free PMC article.
Abstract
Long noncoding RNAs (lncRNAs) regulate gene expression by association with chromatin, but how they target chromatin remains poorly understood. We have used chromatin RNA immunoprecipitation-coupled high-throughput sequencing to identify 276 lncRNAs enriched in repressive chromatin from breast cancer cells. Using one of the chromatin-interacting lncRNAs, MEG3, we explore the mechanisms by which lncRNAs target chromatin. Here we show that MEG3 and EZH2 share common target genes, including the TGF-β pathway genes. Genome-wide mapping of MEG3 binding sites reveals that MEG3 modulates the activity of TGF-β genes by binding to distal regulatory elements. MEG3 binding sites have GA-rich sequences, which guide MEG3 to the chromatin through RNA-DNA triplex formation. We have found that RNA-DNA triplex structures are widespread and are present over the MEG3 binding sites associated with the TGF-β pathway genes. Our findings suggest that RNA-DNA triplex formation could be a general characteristic of target gene recognition by the chromatin-interacting lncRNAs.
Figures
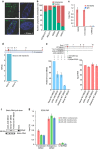
Figure 1. Identification of repressive chromatin-associated lncRNAs using ChRIP-seq.
(a) The ChRIP-seq analysis pipeline used to identify lncRNAs enriched in repressive chromatin. The pie chart shows 276 lncRNAs enriched in both EZH2 and H3K27me3 ChRIP-seq samples compared with the nuclear RNA (input). The P value was obtained by performing a hypergeometric test using all the lncRNAs in our analysis. (b) Bar diagram showing the distribution of T-to-C transitions (indicative of putative RNA–protein contact sites) in input (8,361), EZH2 (18,905) and H3K27me3 (2,651) ChRIP-seq data. Black in the EZH2 bar indicates the number of T-to-C transitions (1,253) that are either present in input or H3K27me3 samples, and blue indicates EZH2-specific T-to-C transitions (17,652). The EZH2-specific T-to-C transitions (17,652) were used to associate with lncRNAs. (c) All the possible conversions present in the EZH2 ChRIP-seq sample. T-to-C conversion and the reverse-strand A-to-G conversions were predominant among all the possible conversion events. (d) LncRNAs (1,046; annotated and non-annotated) harbour EZH2-specific (17,652) T-to-C conversion site. Seventy repressive chromatin-enriched lncRNAs (out of 276) carry T-to-C transitions, including known PRC2-interacting lncRNAs such as MEG3, KCNQ1OT1 and_BDNF-AS1._ The P value was obtained by performing a hypergeometric test using all the lncRNAs considered in our analysis. (e,f) The distribution of the sequencing reads on MEG3 and_KCNQ1OT1_ transcripts from H3K27me3, EZH2-enriched chromatin fractions and input RNA samples. The tags represent the read distribution and the signal represents the intensity of reads over MEG3 and_KCNQ1OT1_ transcripts. Locations of T-to-C transitions over the exons are depicted below the physical maps. The left panel depicts the RPKM (Reads per kilobase per million) for MEG3 and KCNQ1OT1 in H3K27me3, EZH2 ChRIP RNA and input RNA samples. The fold enrichment (FC) in H3K27me3 and EZH2 ChRIP RNA compared with input is indicated. (g) ChRIP validation: RT–qPCR data showing the enrichment of the selected annotated and non-annotated lncRNAs in the EZH2 and H3K27me3 ChRIP pull-downs compared with input. We did not observe any enrichment of these lncRNAs in the H3K4me2 (active chromatin marks) and immunoglobulin G (IgG; nonspecific antibody) ChRIP pull-downs. Actin was used as a negative control. Data represent the mean±s.d. of three independent biological experiments.
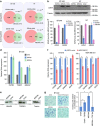
Figure 2. Molecular characterization of MEG3 and PRC2 interaction.
(a) RNA-fluorescence in situ hybridization showing the distribution of the MEG3 signal (green) in the nucleus (blue, stained with 4,6-diamidino-2-phenylindole). An RNase A-treated sample was used as a negative control. Scale bar, 1 μm. (b) RT–qPCR data showing the distribution of lncRNAs and protein-coding RNAs in the nuclear and cytoplasmic fractions (±s.d., n_=3). (c) RT–qPCR analysis of MEG3, KCNQ1OT1 and U1SnRNA in EZH2 RIP-purified RNA from BT-549 cells. U1SnRNA served as negative control. The enrichment is plotted as percentage of input (±s.d.,n_=3). (d) Physical map of the MEG3_containing numbered exons showing two T-to-C transitions. The exons in red are constitutively expressed and blue are alternatively spliced exons. First conversion is part of exon 3 showing higher expression, whereas the second conversion is part of exon 4 showing low expression in the nuclear RNA sequencing. (e) In vitro interaction of MEG3 and PRC2. The schematic indicates the exons of the WT MEG3 clone. Left: RT–qPCR showing enrichment of sense WT MEG3 and MEG3_carrying deletions (Δ340-348 or Δ345-348 MEG3) in_in vitro RNA binding assays. Reaction with antisense WT_MEG3 or without purified PRC2 served as negative controls. The binding efficiency of MEG3 deletions were presented relative to WT_MEG3 (±s.d., n_=3). Right: RT–qPCR showing the quantification of input RNAs. (f) Upper panel: western blot showing EZH2 levels after pull-down with biotinylated sense WT MEG3, antisense WT MEG3, and Δ345-348_MEG3 RNAs incubated with nuclear extract. This is a representative data set from several experiments. Lower panel: agarose gel picture showing input biotin-RNA. (g) RT–qPCR result showing the relative enrichment of WT MEG3, Δ340-348 and Δ345-348 MEG3 RNAs in the EZH2-RIP, performed after BT-549 cells were transfected with WT and mutant MEG3 plasmids. Data were normalized to the input RNAs and plotted as percentage of input (±s.d., n_=3). To distinguish the endogenous_MEG3 from the ectopically expressed MEG3, we designed RT–qPCRs primers, with one primer mapped to the transcribed portion of the vector and the other to MEG3 RNA. Endogenous_MEG3 served as positive control and U1SnRNA as negative control.
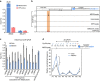
Figure 3. MEG3/EZH2 functional interaction regulates TGF-β pathway genes.
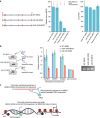
(a–c) MEG3 and EZH2 share common gene targets. (a) Venn diagram showing the number of genes deregulated after downregulation of MEG3 and EZH2 using siRNA in BT-549 and HF cells, and the degree of overlap between the MEG3- and EZH2-dependent genes. The P values were obtained by hypergeometric test using all protein-coding genes as a background. (b) EZH2 protein levels, as determined by western blotting, following EZH2 and MEG3_downregulation in BT-549 and HF cells. Tubulin was used as a loading control. (c) RT–qPCR analysis of EZH2 and_MEG3 mRNA expression in Ctrlsi, _EZH2_si and _MEG3_si transfected BT-549 and HF cells (±s.d., n_=3). (d) RT–qPCR analysis of TGFB2, TGFBR1 and_SMAD2 gene expression in Ctrlsi, _MEG3_si and EZH2_si transfected BT-549 cells (±s.d., n_=3). (e) Immunoblot showing SMAD2, TGFBR1 and tubulin protein levels following transfection of BT-549 cells with Ctrlsi and MEG3_si. (f) Bar graph showing RT–qPCR analysis of TGFB2,TGFBR1 and SMAD2 mRNA levels after overexpression of_MEG3 (pREP4_MEG3) in BT-549 and MDA-MB-231 cells. The levels in pREP4_MEG3 are presented relative to CtrlpREP4 (±s.d., n_=3). EZH2 was used as a control showing no change in expression after overexpression of MEG3. The_P values were calculated using Student's _t_-test (two-tailed, two-sample unequal variance), *P<0.05. (g) Downregulation of MEG3 influences the invasive property of BT-549 cells through regulation of the TGF-β pathway. Images showing Matrigel invasion of the BT-549 cells. The two images in the upper panel show invasion of BT-549 cells infected with Ctrlsh lentivirus, and Ctrlsh infection followed by incubation with TGF-β2 ligand (Ctrlsh-TGFB2). The images in the bottom panel show the cells infected with_MEG3_Sh and _MEG3_sh infection followed by incubation with TGF-β inhibitor (_MEG3_sh+_TGF-β_in). Scale bar, 10 μm. The bar graph shows quantification (±s.d., _n_=3) of the matrix invaded cells in_MEG3_sh relative to the Ctrlsh. The P values were calculated using Student's _t_-test (two-tailed, two-sample unequal variance), *P<0.05, **P<0.01.
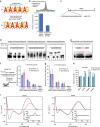
Figure 4. Genome-wide mapping of MEG3 lncRNA binding sites.
(a) RT–qPCR analysis showing specific enrichment (presented as percentage of input) of MEG3 but not MALAT1 RNA in the ChOP pull-down assay with MEG3 antisense probes. The ChOP pull-down with GFP antisense oligo, used as a negative control, did not show any enrichment of MEG3 and MALAT1 RNAs. (b) Genomic tracks showing ChOP-seq (MEG3, GFP and input) and ChIP-seq (H3K4me1) intensities, visualized in log scale. The MEG3 binding site is located upstream of the TGFBR1 gene (falls within the intron of the COL15A1 gene) and it overlaps with H3K4me1 peaks in BT-549 cells. (c) ChIP–qPCR showing enrichment of H3K27me3 chromatin marks, presented as percentage of input, over the MEG3 peaks associated with the TGF-β genes in Ctrlsh and _MEG3_sh cells (±s.d., _n_=3). (d) Schematic outline of the TGFBR1 gene showing MEG3 peaks and the location of 3C primers (P1–P9), as indicated by arrows. EcoRI restriction sites are shown as blue vertical lines. Each error bar represents ±s.d. from three experiments. Looping events between the upstream MEG3 binding site (corresponding to P2 primer) and the TGFBR1 promoter detected by 3C–qPCR in Ctrlsh and _MEG3_sh cells. The P values were calculated using Student's _t_-test (two-tailed, two-sample unequal variance), *P<0.05.
Figure 5. MEG3 lncRNA regulates its target genes through triplex structure formation.
(a) Predicted GA-rich motifs enriched in all MEG3 peaks and peaks associated with deregulated genes using MEME-ChIP. (b) Number of TrTS over the MEG3 peak summits and neighbouring regions, predicted by Triplexator. (c) The schematic shows the MEG3 TFO used in the triplex assays. The exons are colour-coded as described before. (d) Electrophoretic mobility shift assay. End-labelled dsDNA oligos (sequences provided in the schematic with gene name) were incubated alone (lane 1) or with increasing concentrations of_MEG3_ ssRNA TFO (lanes 2 and 3: shift indicated with arrow) or with increasing concentrations of control ssRNA oligo (lanes 8 and 9). dsDNA oligos were incubated with MEG3 TFO and treated with either RNase A (lane 4) or RNase H (lane 5). dsDNA oligos were incubated with MEG3_ssRNA TFO in the presence of either unlabelled specific competitor (lane 6) or nonspecific competitor (lane 7). (e) TGFBR1-associated_MEG3 peak sequence and its mutated version (the changed nucleotides are in red) were incubated alone (lanes 1 and 7) or with_MEG3 TFO. Arrow indicates complex formation. (f,g) Enrichment of MEG3 peak sequences using biotin- and psoralen-labelled_MEG3 TFO. RNase H-treated lysates were used to capture the labelled MEG3 TFO using streptavidin beads. The enrichment of_MEG3_ peaks is presented as the ratio between MEG3 TFO and control oligo (±s.d., _n_=3). (h) RT–qPCR analysis of gene expression in BT-549 cells transfected with either MEG3 TFO or control RNA oligo. Expression in _MEG3_TFO presented relative to the control oligo (±s.d.,n_=3). *P<0.05, Student's_t_-test (two-tailed, two-sample unequal variance). (i) Left panel: CD spectra of a 1:1 mixture of TGFB2 dsDNA and MEG3 TFO (ssRNA) are shown in black, and TGFB2 dsDNA and the control ssRNA are shown in red. Right panel: the sum of the individual CD spectra for_TGFB2 dsDNA and MEG3 TFO (ssRNA) is shown in black, and the sum of the individual CD spectra for TGFB2 dsDNA and the control ssRNA is shown in red. Inset in the left and right panel shows the difference between the two spectra.
Figure 6. RNA–DNA triplexes are present in vivo.
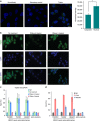
(a) Confocal microscopic images showing immunostaining with anti-triplex dA.2rU antibody (green) in BT-549 cells. The nucleus is stained with DAPI (4,6-diamidino-2-phenylindole; blue). Immunostaining with no antibody and secondary antibody were used as negative controls. Scale bar, 5 μm. The graph to the right shows quantification of the triplex signal in cytoplasm and nuclear compartments obtained from the three-dimensional confocal images. The graph represents the average of cytoplasmic and nuclear signals from >50 cells in several microscopic fields. The error bars indicate s.e.m. The P value was calculated using Student's t_-test **P<0.01. (b) RNA–DNA triplex structures are sensitive to RNase A but are resistant to RNase H in vivo. Top panel: immunofluorescent staining of BT-549 cells with anti-triplex dA.2rU antibody (green) with no treatment (left), pretreated with RNase A (centre), or pretreated with RNase H (right) as indicated. Middle panel: cells were counterstained with DAPI (blue). Bottom panel: overlay of the triplex signals with DAPI staining. Scale bar, 5 μm. (c) Triplex-ChIP–qPCR showing enrichment (presented as percentage of input) of triplex structures over the_MEG3 peaks associated with the TGF-β pathway genes_(TGFBR1_, TGFB2 and SMAD2) in BT-549 cells (±s.d., _n_=3). Actin was used as a negative control. Chromatin was pretreated with RNase A or RNase H before ChIP. Immunoglobulin G (IgG) was used as an antibody control. (d) Triplex-ChIP–qPCR showing enrichment (presented as percentage of input) of triplex structures over the MEG3 peaks associated with the TGF-β pathway genes (TGFBR1, TGFB2 and SMAD2) in Ctrlsh and _MEG3_sh BT-549 cells (±s.d.,_n_=3). IgG was used as an antibody control.
Figure 7. Chromatin-binding sequences and PRC2-binding sequences of MEG3 lncRNA are functionally distinct.
(a) MEG3_-PRC2 in vitro binding assay. Left panel: schematic representation of WT MEG3, Δ46-56_MEG3 and Δ345-348_MEG3_. Green and red boxes indicate PRC2- and chromatin-interacting sequences, respectively. Middle panel: bar diagram showing the relative binding efficiency (as determined by RT–qPCR) of the sense WT MEG3, Δ46-56 MEG3 and Δ345-348 MEG3 RNAs in an in vitro PRC2-binding assay. Binding assays with no PRC2 and antisense WT MEG3 served as negative controls. The PRC2-binding efficiency of sense WT MEG3 was set to 100, and the binding efficiency of the MEG3 mutants is presented relative to WT MEG3 (±s.d.,n_=3). Right panel: RT–qPCR showing the quantification of the input sense WT MEG3, MEG3 deletions (Δ340-348 or Δ345-348 MEG3) and antisense WT MEG3_RNAs. (b) Deletion of MEG3 TFO leads to loss of chromatin interaction. Left panel: schematic display of interaction of the WT_MEG3 and MEG3 mutants (Δ46-56_MEG3 and Δ345-348_MEG3_) with the MEG3 peak sequences in vivo. Red (MEG3 TFO) and green (PRC2-interacting region) colour-coded regions indicate the location of the deleted MEG3 RNA sequences 46-56 and 345-348, respectively. Biotin-labelled WT MEG3 or_MEG3_ mutants were used to transfect BT-549 cells followed by crosslinking with formaldehyde. RNAse H-treated cell lysates were incubated with streptavidin beads to capture the MEG3 RNA-associated DNA. Middle panel: qPCR data are presented as the ratio of captured DNA in WT_MEG3_ or MEG3 mutants to captured non-biotinylated_MEG3_ RNA (±s.d., n_=3). Right panel: agarose gel picture showing the quality of the biotin-labelled WT and mutant_MEG3 RNAs (500 ng of each biotin-RNA was loaded). (c) Model depicting how chromatin-interacting sequences of_MEG3_ lncRNA-containing GA-rich sequences form RNA–DNA triplex with the GA-rich DNA sequences to guide MEG3 lncRNA to chromatin. PRC2-interacting sequences of MEG3 lncRNA facilitate recruitment of the PRC2 to distal regulatory elements, thereby establishing H3K27me3 marks to modulate gene expression.
Similar articles
- MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines.
Terashima M, Tange S, Ishimura A, Suzuki T. Terashima M, et al. J Biol Chem. 2017 Jan 6;292(1):82-99. doi: 10.1074/jbc.M116.750950. Epub 2016 Nov 16. J Biol Chem. 2017. PMID: 27852821 Free PMC article. - Long Noncoding RNA Meg3 Regulates Mafa Expression in Mouse Beta Cells by Inactivating Rad21, Smc3 or Sin3α.
Wang N, Zhu Y, Xie M, Wang L, Jin F, Li Y, Yuan Q, De W. Wang N, et al. Cell Physiol Biochem. 2018;45(5):2031-2043. doi: 10.1159/000487983. Epub 2018 Mar 6. Cell Physiol Biochem. 2018. PMID: 29529600 - Long Noncoding RNA MEG3 Is an Epigenetic Determinant of Oncogenic Signaling in Functional Pancreatic Neuroendocrine Tumor Cells.
Iyer S, Modali SD, Agarwal SK. Iyer S, et al. Mol Cell Biol. 2017 Oct 27;37(22):e00278-17. doi: 10.1128/MCB.00278-17. Print 2017 Nov 15. Mol Cell Biol. 2017. PMID: 28847847 Free PMC article. - RNA-DNA Triplex Formation by Long Noncoding RNAs.
Li Y, Syed J, Sugiyama H. Li Y, et al. Cell Chem Biol. 2016 Nov 17;23(11):1325-1333. doi: 10.1016/j.chembiol.2016.09.011. Epub 2016 Oct 20. Cell Chem Biol. 2016. PMID: 27773629 Review. - Long noncoding RNAs: Novel insights into hepatocelluar carcinoma.
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. He Y, et al. Cancer Lett. 2014 Mar 1;344(1):20-27. doi: 10.1016/j.canlet.2013.10.021. Epub 2013 Oct 30. Cancer Lett. 2014. PMID: 24183851 Review.
Cited by
- Biological relevance and therapeutic potential of G-quadruplex structures in the human noncoding transcriptome.
Tassinari M, Richter SN, Gandellini P. Tassinari M, et al. Nucleic Acids Res. 2021 Apr 19;49(7):3617-3633. doi: 10.1093/nar/gkab127. Nucleic Acids Res. 2021. PMID: 33721024 Free PMC article. Review. - Long noncoding RNAs in cancer: mechanisms of action and technological advancements.
Bartonicek N, Maag JL, Dinger ME. Bartonicek N, et al. Mol Cancer. 2016 May 27;15(1):43. doi: 10.1186/s12943-016-0530-6. Mol Cancer. 2016. PMID: 27233618 Free PMC article. Review. - The long non-coding RNA maternally expressed gene 3 activates p53 and is downregulated in esophageal squamous cell cancer.
Lv D, Sun R, Yu Q, Zhang X. Lv D, et al. Tumour Biol. 2016 Oct 24. doi: 10.1007/s13277-016-5426-y. Online ahead of print. Tumour Biol. 2016. PMID: 27778235 - Comprehensive multi-omics analysis uncovers a group of TGF-β-regulated genes among lncRNA EPR direct transcriptional targets.
Zapparoli E, Briata P, Rossi M, Brondolo L, Bucci G, Gherzi R. Zapparoli E, et al. Nucleic Acids Res. 2020 Sep 18;48(16):9053-9066. doi: 10.1093/nar/gkaa628. Nucleic Acids Res. 2020. PMID: 32756918 Free PMC article. - LncRNA POU3F3 promotes cancer cell migration and invasion in nasopharyngeal carcinoma by up-regulating TGF-β1.
Li W, Wu X, She W. Li W, et al. Biosci Rep. 2019 Jan 25;39(1):BSR20181632. doi: 10.1042/BSR20181632. Print 2019 Jan 31. Biosci Rep. 2019. PMID: 30602452 Free PMC article.
References
- Mohammad F., Mondal T., Guseva N., Pandey G. K. & Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137, 2493–2499 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials