SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells - PubMed (original) (raw)
. 2015 Sep 18;43(16):7931-44.
doi: 10.1093/nar/gkv722. Epub 2015 Jul 22.
Affiliations
- PMID: 26206670
- PMCID: PMC4652757
- DOI: 10.1093/nar/gkv722
SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells
Meryem Alagoz et al. Nucleic Acids Res. 2015.
Abstract
Recent studies have shown that homologous recombination (HR) requires chromatin repression as well as relaxation at DNA double strand breaks (DSBs). HP1 and SUV39H1/2 are repressive factors essential for HR. Here, we identify SETDB1 as an additional compacting factor promoting HR. Depletion of HP1, SUV39, SETDB1 or BRCA1 confer identical phenotypes. The repressive factors, like BRCA1, are dispensable for the initiation of resection but promote the extension step causing diminished RPA or RAD51 foci and HR in irradiated G2 cells. Depletion of the compacting factors does not inhibit BRCA1 recruitment but at 8 h post IR, BRCA1 foci are smaller and aberrantly positioned compared to control cells. BRCA1 promotes 53BP1 repositioning to the periphery of enlarged foci and formation of a devoid core with BRCA1 becoming enlarged and localized internally to 53BP1. Depletion of the compacting factors precludes these changes at irradiation-induced foci. Thus, the repressive factors are required for BRCA1 function in promoting the repositioning of 53BP1 during HR. Additionally, depletion of these repressive factors in undamaged cells causes diminished sister chromatid association at centromeric sequences. We propose a model for how these findings may be functionally linked.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
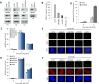
Figure 1.
SETDB1, SUV39 and HP1 are dispensable for DSB repair in G1 phase but essential in G2. (A) 1BR3 hTERT cells were transfected with siRNAs targeting control (CTR), SETDB1, SUV39, HP1α/β and γ and BRCA1. Knockdown of each component was verified by immunoblotting and H3K9me3 levels assessed in whole cell extracts. (B) Quantification of the reduction in H3K9me3 levels. Note that in panels A and B we used combined oligonucleotides for HP1α/β and γ. (C and D) Following siRNA mediated knockdown as indicated, 1BR3 hTERT cells were irradiated with 3 Gy and γH2AX foci enumerated at the indicated times in (C) G1 (CENPF−) and (D) G2 (CENP+) cells. (E and F) Representative images from experiments (C and D). Staining for γH2AX, CENPF and DAPI is as indicated. Asterisks denote statistically significant differences (P < 0.05); _t_-test). Results represent the mean ± S.E.M of 3 experiments. (G) Chromosomal breaks were determined by premature chromosome condensation (PCC) breakage analysis. 1BR3 hTERT cells were subjected to siRNA mediated knockdown of control (CTR) and SETDB1 and irradiated with 2 Gy IR. Chromosomal breaks were assessed at 8 h post IR. Data are represent the mean ± S.E.M of three experiments.
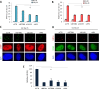
Figure 2.
SETDB1, SUV39 and HP1 are required for the formation of RPA and RAD51 foci and for HR. (A and B) 1BR3 hTERT cells were transfected with siRNAs targeting control (CTR), SETDB1, SUV39, HP1 and BRCA1, irradiated with 3 Gy and 2 h post IR, (A) RPA and (B) RAD51 foci were quantified. RPA and RAD51 analysis at additional time points is shown in Supplementary Figure S1. Asterisks denote statistically significant differences between control (CTR) and SETDB1, SUV39, HP1 knockdown cells. (P < 0.001;t_-test). (C and D) Representative images from the above experiments: (C) RPA and CENPF (D) RAD51 and CENPF as indicated. E. U2OS DR-GFP cells were transfected with siRNAs targeting control (CTR), BRCA1, SETDB1, SUV39 or HP1 and 24 h later were re-transfected with pCBA_SceI. A GFP signal is generated following intrachromosomal gene conversion following I-_SceI-_induced DSB formation. The percentage of GFP+ cells was measured by FACS 48 h after transfection with I-Sce1. GFP+ cells were normalized to the percentage of EGFP positive cells and the level of the S/G2 population. All samples had a similar fraction of G2 phase cells and expression of I-Sce1 was shown to be similar by Western blotting using an HA-tag on I-Sce1 (Supplementary Figure S2A, B). Asterisks denote statistically significant differences (P<0.001; _t_-test). Results represent the mean ± S.E.M of 3 experiments.
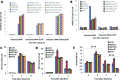
Figure 3.
SETDB1, HP1 and SUV39 function downstream of the initiation of resection. (A) Following transfection of 1BR3 hTERT cells with siRNAs targeting control (CTR), SETDB1, SUV39, HP1 with or without co-depletion with CtIP or EXO1/BLM, cells were irradiated with 3 Gy and γH2AX foci enumerated at the times indicated. Asterisks denote statistically significant differences (_P<_0.01; _t_-test). Knockdown of EXO/BLM is shown in Supplementary Figure S3A. (B) Cells treated as above were enumerated for RAD51 foci at 2 h post IR. (C and D) Following siRNA mediated knockdown as in panel A, cells were exposed to 3 Gy IR and stained with RIF-1 antibodies at the indicated times. RIF1 foci were counted in (C) G1 and (D) G2 cells. (E) The ratio of RIF1/γH2AX foci in G1 versus G2 cells at 2 and 8 h post 3 Gy IR. Data are the mean ± S.E.M of three experiments.
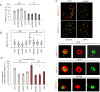
Figure 4.
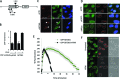
SETDB1, SUV39 and HP1 are dispensable for BRCA1 recruitment but required for its enlargement and localization within IRIF. (A) BRCA1 foci were enumerated in 1BR3 hTERT cells at 0.5 and 8 h in G2 cells post 3 Gy IR following the indicated siRNA-mediated knockdown. Note that Delta Vision microscopy was used for this analysis in contrast to Figure 1. The number of foci (γH2AX and BRCA1) is elevated when enumerated using Delta Vision microscopy due to the better delivery of the light source to the objective lens and enhanced contrast. Thus, foci numbers here do not correlate with those in Figure 1 although the relative impact of siRNA remains consistent. (B) BRCA1 foci size was analysed in G2 cells after 3 Gy IR at the indicated time points using softWoRx® suite software. Data are the mean ± S.E.M of three experiments. Asterisks denote statistically significant differences (P < 0.05; _t_-test). (C) 1BR3 hTERT cells were transfected with siRNAs targeting control (CTR) or SETDB1, SUV39, HP1, 48 h later cells were irradiated with 3 Gy and co-stained with BRCA1 and 53BP1 antibody at 8 h post IR. Images were taken by Delta Vision microscopy. In the bottom panel, representative 3D images were generated by the softWoRx® suite software. Panels shows representative images of 53BP1 and/or BRCA1 at 8 h post IR. Top two lines of the bottom panel show two images after siCTR and the bottom row after siRNA SETDB1 (siSETDB1). Images highlight lack of overlap of 53BP1 and BRCA1 foci after knockdown of SETDB1 and the lack of a devoid core. Knockdown of the other compacting factors gave images similar to those after siRNA SETDB1. Note that the top images show a 2D representation and not all foci are therefore observed. (D) The overlap of BRCA1 and 53BP1 foci was quantified in deconvolved Delta Vision 2D images by the softWoRx® suite software. The result indicates percentage of overlapping foci between (BRCA1+ 53BP1) relative to total BRCA1 foci. The Pearson coefficient of correlation monitors how closely two intensities overlap on a pixel-by-pixel basis (full overlapping is 1.0). Asterisks indicate statistical difference (P < 0.05; _t_-test).
Figure 5.
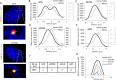
SETDB1, SUV39 and HP1 are required for the repositioning of 53BP1 to the periphery of IRIF during HR. (A) Representative images of 53BP1 (red) and BRCA1 (green) foci used to generate the analysis in panels A–D. This figure shows how a line is drawn through the foci. Following siRNA control, 53BP1 entirely surrounds BRCA1 whereas following siRNA SETDB1 (siSETDB1) the two foci are off set. (B) A549 cells were depleted with siRNAs targeting control (CTR), SETDB1, SUV39, HP1, irradiated with 3 Gy IR and 48 h later co-stained with BRCA1 (green) and 53BP1 (red). The fluorescence intensity was quantified along a line drawn through the centre of 53BP1 foci to provide a fluorescence intensity profile using the softWoRx® suite software. In control cells 53BP1 has a bimodal distribution with BRCA1 being localized within the core of diminished intensity. (C, D and E) Similar quantification was carried out following siRNAs targeting control (CTR), SETDB1, SUV39 and HP1. The results show a monomodal distribution of 53BP1. The width of the peak for 53BP1 and BRCA1 are smaller than in the siCTR sample and the BRCA1 peak is off set relative to the 53BP1 peak. (F) Quantification of the width of 53BP1 foci. The foci width was determined by measuring the distance between the outer edges of the peaks at 50% intensity. (G) BRCA1 intensity following siRNA SETDB1, SUV39 or HP1 was reduced compared to control siRNA. In panels A–D, the maximum Y axis value was automatically set to the maximum fluorescence intensity of 53BP1 (left axis) and BRCA1 (right axis) giving the same size peak irrespective of absolute intensity. Note that BRCA1 intensity is about 10-fold less than the 53BP1 intensity. This panel shows the comparative intensity of BRCA1 following siRNA targeting control (CTR), SETDB1, SUV39 or HP1. Asterisks shows statistically significant difference between control and siRNA SETDB1, SUV39 or HP1 (P < 0.005; _t_-test).
Figure 6.
SETDB1 is recruited to DSB sites. (A) The pCBA_Sce1_ plasmid used to assess protein localization. F1 and F2 represent a primer pair used in panel B. The primers are located 500 bp upstream of the I-SceI recognition site. (B) HeLa DR-GFP cells were transfected with or without pCBASceI plasmid and processed for the ChIP assay using α—yH2AX and α-SETDB1 antibodies at 24 h post-transfection. DNA was recovered from immunoprecipitates and quantified by real time quantitative PCR. (C) U2OS cells were transfected with siRNA targeting control (CTR) or SETDB1. Cells were pre-photosensitized with 10 pM BrdU for 48 h and irradiated with a 405 nm laser. Twenty minutes post-micro irradiation, cells were fixed after pre-extraction and stained for γH2AX, SETDB1 and DAPI. (D) U2OS cells were transfected with siRNAs targeting control (CTR), p150CAF-1 (CAF-1) or HP1α and 48 h later, cells were microirradiated as in C. Twenty minutes later, cells were pre-extracted, fixed and stained for SETDB1, γH2AXor DAPI. (E) U2OS cells transfected with the pEGFP-SETDB1. After 48 h transfection cells were irradiated with a 405 nm laser. A total of 10 μM ATM inhibitor (KU-55933) was added to the cells 1 h prior to microirradiation. The GFP signal intensity was quantified along the laser track in untreated and ATMi-treated cells expressing pEGFP-SETDB1. GFP-signal intensity was monitored in 40 cells for 35 min post IR. Results represent the mean ± S.E.M of three experiments. (F) U2OS cells were treated with 10 μM ATM inhibitor (KU-55933) for 1 h prior to microirradiation as in panel C. Twenty minutes later, cells were processed for immunofluorescence using α-SETBD1 or α—yH2AX antibodies. Reduced γ-H2AX formation and SETDB1 accumulation was observed in cells treated with ATMi compared to control cells.
Figure 7.
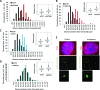
Depletion of HP1, SUV39 or SETDB1 impairs sister chromatid association. (A, B, C and D) The distance between sister chromatids was quantified in 1BR3 hTERT G2 cells transfected with siRNAs targeting control (CTR), SETDB1 (A), SUV39 (B), HP1 (C) and BRCA1 (D) using IF-FISH assay. The SoftWoRx® suite software was used to measure distance between the sister chromatids. The distance between the outer edges of the foci were measured. The distances between signals were measured from three independent experiments and the distribution was plotted as a histogram. Each individual experiment gave a similar difference in distribution. The box plot represents the corresponding clustered columns. Distribution counts from the FISH experiments were analysed using Mann–Whitney Rank Sum Test. (E) IF-FISH images for control siRNA and siRNA SETDB1. Not that since then two FISH spots frequently overlap, the distance between the outer edges were measured. The distances do not, therefore, represent actual sister separation differences but provide a readout for sister association.
Similar articles
- HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response.
Lee YH, Kuo CY, Stark JM, Shih HM, Ann DK. Lee YH, et al. Nucleic Acids Res. 2013 Jun;41(11):5784-98. doi: 10.1093/nar/gkt231. Epub 2013 Apr 15. Nucleic Acids Res. 2013. PMID: 23589625 Free PMC article. - Opposing roles for 53BP1 during homologous recombination.
Kakarougkas A, Ismail A, Klement K, Goodarzi AA, Conrad S, Freire R, Shibata A, Lobrich M, Jeggo PA. Kakarougkas A, et al. Nucleic Acids Res. 2013 Nov;41(21):9719-31. doi: 10.1093/nar/gkt729. Epub 2013 Aug 22. Nucleic Acids Res. 2013. PMID: 23969417 Free PMC article. - Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection.
Kakarougkas A, Ismail A, Katsuki Y, Freire R, Shibata A, Jeggo PA. Kakarougkas A, et al. Nucleic Acids Res. 2013 Dec;41(22):10298-311. doi: 10.1093/nar/gkt802. Epub 2013 Sep 5. Nucleic Acids Res. 2013. PMID: 24013561 Free PMC article. - DNA DSB repair pathway choice: an orchestrated handover mechanism.
Kakarougkas A, Jeggo PA. Kakarougkas A, et al. Br J Radiol. 2014 Mar;87(1035):20130685. doi: 10.1259/bjr.20130685. Br J Radiol. 2014. PMID: 24363387 Free PMC article. Review. - Regulation of repair pathway choice at two-ended DNA double-strand breaks.
Shibata A. Shibata A. Mutat Res. 2017 Oct;803-805:51-55. doi: 10.1016/j.mrfmmm.2017.07.011. Epub 2017 Jul 29. Mutat Res. 2017. PMID: 28781144 Review.
Cited by
- The human HELLS chromatin remodelling protein promotes end resection to facilitate homologous recombination and contributes to DSB repair within heterochromatin.
Kollárovič G, Topping CE, Shaw EP, Chambers AL. Kollárovič G, et al. Nucleic Acids Res. 2020 Feb 28;48(4):1872-1885. doi: 10.1093/nar/gkz1146. Nucleic Acids Res. 2020. PMID: 31802118 Free PMC article. - Role of Histone Methylation in Maintenance of Genome Integrity.
Mushtaq A, Mir US, Hunt CR, Pandita S, Tantray WW, Bhat A, Pandita RK, Altaf M, Pandita TK. Mushtaq A, et al. Genes (Basel). 2021 Jun 29;12(7):1000. doi: 10.3390/genes12071000. Genes (Basel). 2021. PMID: 34209979 Free PMC article. Review. - A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress.
McMurchy AN, Stempor P, Gaarenstroom T, Wysolmerski B, Dong Y, Aussianikava D, Appert A, Huang N, Kolasinska-Zwierz P, Sapetschnig A, Miska EA, Ahringer J. McMurchy AN, et al. Elife. 2017 Mar 15;6:e21666. doi: 10.7554/eLife.21666. Elife. 2017. PMID: 28294943 Free PMC article. - Timely double-strand break repair and pathway choice in pericentromeric heterochromatin depend on the histone demethylase dKDM4A.
Janssen A, Colmenares SU, Lee T, Karpen GH. Janssen A, et al. Genes Dev. 2019 Jan 1;33(1-2):103-115. doi: 10.1101/gad.317537.118. Epub 2018 Dec 21. Genes Dev. 2019. PMID: 30578303 Free PMC article. - Encounters in Three Dimensions: How Nuclear Topology Shapes Genome Integrity.
Sebastian R, Aladjem MI, Oberdoerffer P. Sebastian R, et al. Front Genet. 2021 Oct 21;12:746380. doi: 10.3389/fgene.2021.746380. eCollection 2021. Front Genet. 2021. PMID: 34745220 Free PMC article. Review.
References
- Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous