The Structure of Immature Virus-Like Rous Sarcoma Virus Gag Particles Reveals a Structural Role for the p10 Domain in Assembly - PubMed (original) (raw)
The Structure of Immature Virus-Like Rous Sarcoma Virus Gag Particles Reveals a Structural Role for the p10 Domain in Assembly
Florian K M Schur et al. J Virol. 2015 Oct.
Abstract
The polyprotein Gag is the primary structural component of retroviruses. Gag consists of independently folded domains connected by flexible linkers. Interactions between the conserved capsid (CA) domains of Gag mediate formation of hexameric protein lattices that drive assembly of immature virus particles. Proteolytic cleavage of Gag by the viral protease (PR) is required for maturation of retroviruses from an immature form into an infectious form. Within the assembled Gag lattices of HIV-1 and Mason-Pfizer monkey virus (M-PMV), the C-terminal domain of CA adopts similar quaternary arrangements, while the N-terminal domain of CA is packed in very different manners. Here, we have used cryo-electron tomography and subtomogram averaging to study in vitro-assembled, immature virus-like Rous sarcoma virus (RSV) Gag particles and have determined the structure of CA and the surrounding regions to a resolution of ∼8 Å. We found that the C-terminal domain of RSV CA is arranged similarly to HIV-1 and M-PMV, whereas the N-terminal domain of CA adopts a novel arrangement in which the upstream p10 domain folds back into the CA lattice. In this position the cleavage site between CA and p10 appears to be inaccessible to PR. Below CA, an extended density is consistent with the presence of a six-helix bundle formed by the spacer-peptide region. We have also assessed the affect of lattice assembly on proteolytic processing by exogenous PR. The cleavage between p10 and CA is indeed inhibited in the assembled lattice, a finding consistent with structural regulation of proteolytic maturation.
Importance: Retroviruses first assemble into immature virus particles, requiring interactions between Gag proteins that form a protein layer under the viral membrane. Subsequently, Gag is cleaved by the viral protease enzyme into separate domains, leading to rearrangement of the virus into its infectious form. It is important to understand how Gag is arranged within immature retroviruses, in order to understand how virus assembly occurs, and how maturation takes place. We used the techniques cryo-electron tomography and subtomogram averaging to obtain a detailed structural picture of the CA domains in immature assembled Rous sarcoma virus Gag particles. We found that part of Gag next to CA, called p10, folds back and interacts with CA when Gag assembles. This arrangement is different from that seen in HIV-1 and Mason-Pfizer monkey virus, illustrating further structural diversity of retroviral structures. The structure provides new information on how the virus assembles and undergoes maturation.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
FIG 1
In vitro assembly and purification of VLPs. (a) Diagram of RSV Gag (top) and the extent of the GagΔMBD construct (black rectangle, bottom). GagΔMBD starts at residue 84 of MA. (b) Sedimentation purification of RSV GagΔMBD VLPs. VLPs banded to 40 to 50% (wt/wt) sucrose were separated from unassembled and aggregated protein on a 30 to 60% sucrose step gradient. (c) Negative stain EM image of purified VLPs at low (left) and high (right) magnification (the scale bars are 500 and 100 nm, respectively).
FIG 2
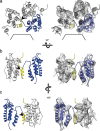
Structure of the immature RSV p10-CA-SP lattice. (a) Sum of 10 computational slices through a Gaussian-filtered tomogram containing one RSV GagΔMBD VLP. The protein density is black. The red box marks the approximate radial position of the capsid density. Scale bar, 25 nm. (b to d) Isosurface representations of the final subtomogram average of the immature RSV p10-CA-SP lattice as derived from subtomogram averaging. High-resolution structures for p10 (yellow), CA-NTD (blue), and CA-CTD (red) have been flexibly fitted into the density. The density is shown as a side view in panel b, from the outside of the VLP in panel c, and from the inside of the VLP in panel d. Scale bars, 25 Å. See also Movie S1 in the supplemental material. (e) Views of the final structural model of the immature RSV p10-CA-SP lattice from outside the virus showing the indicated CA domains. The coloring is as described for panels b to d. Six- and three-fold symmetry axes are indicated by black hexagons and triangles, respectively.
FIG 3
Interactions and structural details in the immature RSV lattice. (a) Flexible fit of RSV-CA into the EM density. Helices are numbered. An arrow indicates the position at which contacts are formed between the helices 9 in the vicinity of Pro426/Val427. The density for the helix-7/helix-8 linker is visible. (b) The CA-CTD forms interactions around the hexameric ring. Residues in the MHR (green) contribute to these interfaces. The putative 6-helix bundle (45) (purple) has been fitted into the density. (c) Isosurface representation of the density corresponding to RSV-SP shown in a side view of the VLP, with the six-helix bundle model generated for RSV-SP (45) fitted into the density. The dashed line indicates the approximate position of the cleavage site between CA and SP.
FIG 4
p10 is an integral part of the assembled lattice. Representations of p10-CA-NTD interactions are shown. Flexible fits alone and with the corresponding EM density are shown in the left and right panels, respectively. Arrowheads mark the proteolytic cleavage site between p10 and CA. (a) Three p10-CA-NTD monomers are shown from the outside of the VLP with one monomer highlighted in yellow (p10) and blue (CA-NTD). The hole on the 6-fold axis is indicated by black lines. (b and c) Side view of two p10-CA-NTD monomers as seen from the hexamer center (b) and rotated by 180° around the vertical axis (c). For clarity, one p10-CA-NTD monomer has been removed from the side views in panels b and c.
FIG 5
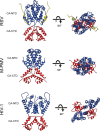
Comparison of immature capsid assembly in different retroviruses. A comparison of immature retroviral capsid dimers (RSV, M-PMV, and HIV-1) is presented. Dimers are shown in orthogonal and top views. CA-CTD interactions are largely conserved between the retroviruses, while the arrangement of the CA-NTD varies. The coloring is as in Fig. 2.
FIG 6
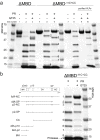
Cleavage of RSV Gag protein and VLPs. (a) Coomassie blue-stained, 5 to 15% gradient gel showing cleavage of unassembled GagΔMBD, unassembled GagΔMBD-11C (E227C and T259C), and purified GagΔMBD-11C (E227C and T259C) VLPs. The lanes represent protein with or without GT25 oligonucleotide (+/−) and with or without PR (+/−). The numbers in lanes 5 and 6 correspond to identification of products in panel b. (b) Lanes 5 and 6 were copied from panel a, with each band annotated to indicate the corresponding cleavage product (see Materials and Methods). Band 9 is protease.
Similar articles
- Maturation of retroviruses.
Pornillos O, Ganser-Pornillos BK. Pornillos O, et al. Curr Opin Virol. 2019 Jun;36:47-55. doi: 10.1016/j.coviro.2019.05.004. Epub 2019 Jun 8. Curr Opin Virol. 2019. PMID: 31185449 Free PMC article. Review. - Role of Mason-Pfizer monkey virus CA-NC spacer peptide-like domain in assembly of immature particles.
Strohalmová-Bohmová K, Spiwok V, Lepšík M, Hadravová R, Křížová I, Ulbrich P, Pichová I, Bednárová L, Ruml T, Rumlová M. Strohalmová-Bohmová K, et al. J Virol. 2014 Dec;88(24):14148-60. doi: 10.1128/JVI.02286-14. Epub 2014 Oct 1. J Virol. 2014. PMID: 25275119 Free PMC article. - Conserved and variable features of Gag structure and arrangement in immature retrovirus particles.
de Marco A, Davey NE, Ulbrich P, Phillips JM, Lux V, Riches JD, Fuzik T, Ruml T, Kräusslich HG, Vogt VM, Briggs JA. de Marco A, et al. J Virol. 2010 Nov;84(22):11729-36. doi: 10.1128/JVI.01423-10. Epub 2010 Sep 1. J Virol. 2010. PMID: 20810738 Free PMC article. - Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution.
Schur FK, Hagen WJ, Rumlová M, Ruml T, Müller B, Kräusslich HG, Briggs JA. Schur FK, et al. Nature. 2015 Jan 22;517(7535):505-8. doi: 10.1038/nature13838. Epub 2014 Nov 2. Nature. 2015. PMID: 25363765 - Assembly and architecture of HIV.
Ganser-Pornillos BK, Yeager M, Pornillos O. Ganser-Pornillos BK, et al. Adv Exp Med Biol. 2012;726:441-65. doi: 10.1007/978-1-4614-0980-9_20. Adv Exp Med Biol. 2012. PMID: 22297526 Free PMC article. Review.
Cited by
- Computational Methodologies for Real-Space Structural Refinement of Large Macromolecular Complexes.
Goh BC, Hadden JA, Bernardi RC, Singharoy A, McGreevy R, Rudack T, Cassidy CK, Schulten K. Goh BC, et al. Annu Rev Biophys. 2016 Jul 5;45:253-78. doi: 10.1146/annurev-biophys-062215-011113. Epub 2016 May 2. Annu Rev Biophys. 2016. PMID: 27145875 Free PMC article. Review. - Biochemical Reconstitution of HIV-1 Assembly and Maturation.
Kucharska I, Ding P, Zadrozny KK, Dick RA, Summers MF, Ganser-Pornillos BK, Pornillos O. Kucharska I, et al. J Virol. 2020 Feb 14;94(5):e01844-19. doi: 10.1128/JVI.01844-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31801870 Free PMC article. - Structure and architecture of immature and mature murine leukemia virus capsids.
Qu K, Glass B, Doležal M, Schur FKM, Murciano B, Rein A, Rumlová M, Ruml T, Kräusslich HG, Briggs JAG. Qu K, et al. Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):E11751-E11760. doi: 10.1073/pnas.1811580115. Epub 2018 Nov 26. Proc Natl Acad Sci U S A. 2018. PMID: 30478053 Free PMC article. - Maturation of retroviruses.
Pornillos O, Ganser-Pornillos BK. Pornillos O, et al. Curr Opin Virol. 2019 Jun;36:47-55. doi: 10.1016/j.coviro.2019.05.004. Epub 2019 Jun 8. Curr Opin Virol. 2019. PMID: 31185449 Free PMC article. Review. - Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Dick RA, Xu C, Morado DR, Kravchuk V, Ricana CL, Lyddon TD, Broad AM, Feathers JR, Johnson MC, Vogt VM, Perilla JR, Briggs JAG, Schur FKM. Dick RA, et al. PLoS Pathog. 2020 Jan 27;16(1):e1008277. doi: 10.1371/journal.ppat.1008277. eCollection 2020 Jan. PLoS Pathog. 2020. PMID: 31986188 Free PMC article.
References
- Macek P, Chmelik J, Krizova I, Kaderavek P, Padrta P, Zidek L, Wildova M, Hadravova R, Chaloupkova R, Pichova I, Ruml T, Rumlova M, Sklenar V. 2009. NMR structure of the N-terminal domain of capsid protein from the Mason-Pfizer monkey virus. J Mol Biol 392:100–114. doi:10.1016/j.jmb.2009.06.029. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials