METABOLISM. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction - PubMed (original) (raw)
METABOLISM. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction
Ling Yang et al. Science. 2015.
Abstract
The association between inflammation and endoplasmic reticulum (ER) stress has been observed in many diseases. However, if and how chronic inflammation regulates the unfolded protein response (UPR) and alters ER homeostasis in general, or in the context of chronic disease, remains unknown. Here, we show that, in the setting of obesity, inflammatory input through increased inducible nitric oxide synthase (iNOS) activity causes S-nitrosylation of a key UPR regulator, IRE1α, which leads to a progressive decline in hepatic IRE1α-mediated XBP1 splicing activity in both genetic (ob/ob) and dietary (high-fat diet-induced) models of obesity. Finally, in obese mice with liver-specific IRE1α deficiency, reconstitution of IRE1α expression with a nitrosylation-resistant variant restored IRE1α-mediated XBP1 splicing and improved glucose homeostasis in vivo. Taken together, these data describe a mechanism by which inflammatory pathways compromise UPR function through iNOS-mediated S-nitrosylation of IRE1α, which contributes to defective IRE1α activity, impaired ER function, and prolonged ER stress in obesity.
Copyright © 2015, American Association for the Advancement of Science.
Figures
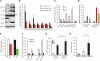
Fig. 1. Inflammation contributes to defective IRE1α-mediated XBP1 splicing in obese liver
(A) UPR status was examined in livers of 16-week-old obese (ob/ob) and lean mice by using phosphorylation of IRE1α, PERK, and JNK, as well as expression of sXBP1 and uXBP1 as markers. Data are representative of results from two to three independent cohorts of mice. (B) Expression of hepatic UPR modulators in 7-, 12- and 16-week-old ob/ob mice relative to lean controls (dashed line indicates lean level), n = 4. (C) ChIP assay examining XBP1 target gene occupancy in primary hepatocytes from 16-week-old ob/ob and lean mice. Data were normalized to 2% input, followed by comparison to each immunoglobulin G (IgG) control (n = 4). Data are representative of results from two independent sets of mice. Asterisk (*) indicates statistical significance between lean and obese mice in (B) and (C). Statistical analysis was performed by multiple t tests and significance was determined by the Holm-Šídák method using Prism (B) and Student's t test (C). AU, arbitrary units. (D) sXBP1 was examined in the livers from lean (RD) and obese (HFD-fed) mice injected with vehicle (V), or tunicamycin (TN, 6 hours, 0.5 mg/kg per kg body weight). sXBP1 expression was also examined in the livers from control mice fed RD or HFD transduced with full-length XBP1 (RD-XBP1, HFD-XBP1) or lean controls (RD-XBP1). Asterisk (*) indicates statistical significance between treatments within the control group, and # indicates statistical significance between RD and HFD [one-way analysis of variance (ANOVA) followed by post hoc Tukey's test], n = 6 to 8 mice. (E) In vitro splicing assays measuring the XBP1 splicing efficiency using hepatic IRE1α from mice with dietary (HFD) and genetic (ob/ob) obesity and controls. sXBP1 expression was examined by qRT-PCR. Results are normalized to lean samples. Asterisk (*) indicates statistical significance between lean and obese mice (Student's t test, n = 3). Data are representative of results from two independent sets of mice. (F) iNOS and eNOS mRNAs were examined in livers of 7- and 16-week-old ob/ob or HFD-fed mice and lean controls by qRT-PCR. Asterisk (*) indicates statistical significance between lean and obese mice (Student's t test), n = 4 to 6. (G) sXBP1 expression was examined by qRT-PCR in primary hepatocytes from lean mice transduced with Ad-shiNOS (iNOS.sh) or control virus (LacZ.sh) followed by treatment with thapsigargin (Tg+) for 2 hours, n = 4. (H) In vitro XBP1 splicing assay using IRE1α purified from the livers ob/ob mice after iNOS suppression (normalized to IgG control). Asterisk (*) indicates statistical significance between treatments and controls, and # indicates statistical significance between iNOS.sh group and LacZ.sh group (one-way ANOVA followed by post hoc Tukey's test). All data are shown as means ± SEM. Data are representative of results from two independent sets of mice. *P < 0.05; #P < 0.05.
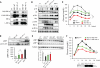
Fig. 2. S-Nitrosylation of IRE1α results in decreased IRE1α RNase activity
(A) General SNO profile in livers of ob/ob mice (obese) versus lean controls. The S-nitrosylated proteins were purified by a biotin-switch method. SNAP was used as a positive (+) control, and a sample without ascorbic acid treatment served as a negative (−) control. Data are representative of results from four independent cohorts of mice. (B) Specific SNO proteins in the livers of obese (ob/ob) and lean mice. The S-nitrosylated proteins were purified by a biotin-switch method and detected by Western blot analysis. (C) In vitro XBP1 splicing assay in IRE1α−/− MEFs reconstituted with IRE1α. The IRE1α protein levels are shown in the gel (top). NMMA is an iNOS inhibitor. AU, arbitrary units. Asterisk (*) indicates statistical significance compared with control, and # indicates statistically significant differences between treatments (one-way ANOVA followed by post hoc Tukey's test). Data are representative of three individual experiments. (D) IRE1α−/− MEFs reconstituted with IRE1α in the presence or absence of iNOS were treated with 10 ng/ml TNFα overnight. A biotin switch was performed, and samples were blotted with the FLAG-specific antibody. Data are representative of two individual experiments. (E) MS analysis of S-nitrosylated peptide 1, which includes the C931 in the RNase domain of human IRE1α. Shown is unmodified peptide 1, single-charged M+ (m/z 1718.8490), and the peptide after GSNO modification (M+NO)+ (m/z 1747.8405). Nitrosylation of peptide 1 results in a m/z change of +28.9915. (Top) A scheme of IRE1α protein structure is shown. (F) qRT-PCR for sXBP1 in IRE1α-deficient primary hepatocytes reconstituted with WT IRE1α, or nitrosylation-resistant IRE1α (IRE1-M1+2) with mutations in two SNO sites in the RNase domain, or IRE1α with a mutation in its RNase domain (IRE1-RD). The cells were treated with 10 ng/ml TNFα overnight in the presence or absence of Tg (100 nM, 2 hours). Results are presented as sXBP1/IRE1α expression levels in each group normalized to controls. (Top) The IRE1α protein levels are shown. All data are shown as means ± SEM. Asterisk (*) indicates statistical significance compared with control, and # indicates statistically significant differences between treatments (one-way ANOVA followed by post hoc Tukey's test, n = 4). *P < 0.05; #P < 0.05.
Fig. 3. S-Nitrosylation of IRE1α RNAse domain results in impaired XBP1 processing
(A) Nitrosylation on C931 and C951 was modeled onto the crystal structure of human IRE1α (PDB 3P23). (B) Urea polyacrylamide gel electrophoresis of sXBP1 substrate (labeled with FAM at the 5′ end) cleaved by different doses of IRE1c in the presence or absence of the chemical NO donor, SNAP (5 mM). RNase A was used as a control. The quantification of cleaved RNA is shown as band intensity (INTmm2). Data are representative of three individual experiments. (C) CLIP assay detecting interaction between IRE1α and unspliced XBP1. IRE1α−/− MEFs were reconstituted with WT IRE1α, followed by treatment with Tg in the absence or presence of TNFα. After UV cross-linking, the IRE1α complexes were immunoprecipitated, and unspliced XBP1 was detected by qRT-PCR. Data are presented as means ± SEM, with n = 6. Data are representative of four individual experiments. (D) Binding of an RNase-resistant analog of XBP1 RNA, HP21 (FAM-dCdCdGdCdAdG) with IRE1c in the absence or presence of NO donor (35) was analyzed by fluorescence polarization assay using a nonfluorescent HP21 as a competitor. The data are presented as units of millipolarization (mP), and the inhibition constant (_K_i) is shown (bottom). Data are representative of three individual experiments. (E) IRE1c oligomerization in the absence or presence of NO donors, SNAP (5 mM) or GSNO (0.25 mM). Data are representative of three individual experiments. *P < 0.05; #P < 0.05.
Fig. 4. Effects of IRE1αa S-nitrosylation on insulin action and glucose homeostasis
(A to D) ob/ob mice were transduced with adenovirus containing sXBP1, WT IRE1α or nitrosylation-resistant IRE1-M1+2. (A) S-Nitrosylation of IRE1α in primary hepatocytes from these mice. The nitrosylation state of IRE1α was examined by biotin switch assay. L, longer exposure time. Data are representative of two individual mouse cohorts. (B) Hepatic insulin action and UPR status. N = 6 to 7, data are representative of two individual cohorts of mice. (C) Glucose tolerance test. All data are presented as means ± SEM, with statistical analysis of the area under the curve (AUC) performed by two-way ANOVA with post hoc Bonferroni test. Asterisk (*) indicates statistical significance compared with control, and # indicates differences between sXBP1 and IRE1-M1+2 expressing mice. Data are representative of two individual cohorts of mice [n = 8 (10-week-old mice)]. (D) sXBP1 expression in the nuclear fraction of liver, with densitometric analysis shown at the bottom of panel. Asterisk (*) indicates statistical significance compared with control, and # indicates statistically significant differences between IRE1-WTand IRE1-M1+2 (one-way ANOVA followed by post hoc Tukey's test). Data are representative of two individual mouse cohorts. (E and F) IRE1α-floxed mice were transduced with Ad-LacZ (Control) or Ad-Cre to delete IRE1α (IRE1LKO), followed by expression of IRE1-WT or the IRE1-M1+2 variant. (E) Hepatic expression of sXBP1 and uXBP1 in the liverof these mice. Quantification of sXBP1/tubulin ratio is shown below. Asterisk (*) indicates statistical significance compared with control, and # indicates statistically significant differences between IRE1-WT and IRE1-M1+2 (one-way ANOVA followed by post hoc Tukey's test). Data are representative of two independent sets of mice. (F) Glucose tolerance test of the mice shown in (E). (Bottom) The IRE1α expression levels are shown [n = 8 (9-week old mice)]. Data are presented as means ± SEM, with statistical analysis of AUC performed by two-way ANOVA with post hoc Bonferroni test. Number sign (#) indicates statistically significant differences comparing IRE1LKO+IRE-M1+2 with IRE1LKO+IRE1-WT, as well as IRE1LKO+IRE1-WT with IRE1LKO. Data are representative of two individual cohorts of mice. *P < 0.05; #P < 0.05.
Similar articles
- Cytoplasmic IRE1alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress.
Back SH, Lee K, Vink E, Kaufman RJ. Back SH, et al. J Biol Chem. 2006 Jul 7;281(27):18691-706. doi: 10.1074/jbc.M602030200. Epub 2006 Apr 27. J Biol Chem. 2006. PMID: 16644724 - ER stress and distinct outputs of the IRE1α RNase control proliferation and senescence in response to oncogenic Ras.
Blazanin N, Son J, Craig-Lucas AB, John CL, Breech KJ, Podolsky MA, Glick AB. Blazanin N, et al. Proc Natl Acad Sci U S A. 2017 Sep 12;114(37):9900-9905. doi: 10.1073/pnas.1701757114. Epub 2017 Aug 28. Proc Natl Acad Sci U S A. 2017. PMID: 28847931 Free PMC article. - Interplay between the oxidoreductase PDIA6 and microRNA-322 controls the response to disrupted endoplasmic reticulum calcium homeostasis.
Groenendyk J, Peng Z, Dudek E, Fan X, Mizianty MJ, Dufey E, Urra H, Sepulveda D, Rojas-Rivera D, Lim Y, Kim DH, Baretta K, Srikanth S, Gwack Y, Ahnn J, Kaufman RJ, Lee SK, Hetz C, Kurgan L, Michalak M. Groenendyk J, et al. Sci Signal. 2014 Jun 10;7(329):ra54. doi: 10.1126/scisignal.2004983. Sci Signal. 2014. PMID: 24917591 Free PMC article. - Stressed out about obesity: IRE1α-XBP1 in metabolic disorders.
Sha H, He Y, Yang L, Qi L. Sha H, et al. Trends Endocrinol Metab. 2011 Sep;22(9):374-81. doi: 10.1016/j.tem.2011.05.002. Epub 2011 Jun 22. Trends Endocrinol Metab. 2011. PMID: 21703863 Free PMC article. Review. - Targeting the IRE1α-XBP1 branch of the unfolded protein response in human diseases.
Jiang D, Niwa M, Koong AC. Jiang D, et al. Semin Cancer Biol. 2015 Aug;33:48-56. doi: 10.1016/j.semcancer.2015.04.010. Epub 2015 May 16. Semin Cancer Biol. 2015. PMID: 25986851 Free PMC article. Review.
Cited by
- The role of nonmyocardial cells in the development of diabetic cardiomyopathy and the protective effects of FGF21: a current understanding.
Zhang T, Jiang D, Zhang X, Chen L, Jiang J, Zhang C, Li S, Li Q. Zhang T, et al. Cell Commun Signal. 2024 Sep 26;22(1):446. doi: 10.1186/s12964-024-01842-0. Cell Commun Signal. 2024. PMID: 39327594 Free PMC article. Review. - Mechanisms of obesity- and diabetes mellitus-related pancreatic carcinogenesis: a comprehensive and systematic review.
Ruze R, Song J, Yin X, Chen Y, Xu R, Wang C, Zhao Y. Ruze R, et al. Signal Transduct Target Ther. 2023 Mar 24;8(1):139. doi: 10.1038/s41392-023-01376-w. Signal Transduct Target Ther. 2023. PMID: 36964133 Free PMC article. - The therapeutic potential of second and third generation CB1R antagonists.
Cinar R, Iyer MR, Kunos G. Cinar R, et al. Pharmacol Ther. 2020 Apr;208:107477. doi: 10.1016/j.pharmthera.2020.107477. Epub 2020 Jan 9. Pharmacol Ther. 2020. PMID: 31926199 Free PMC article. Review. - GSNOR negatively regulates the NLRP3 inflammasome via S-nitrosation of MAPK14.
Liu Q, Jiao L, Ye MS, Ma Z, Yu J, Su LY, Zou WY, Yang LX, Chen C, Yao YG. Liu Q, et al. Cell Mol Immunol. 2024 Jun;21(6):561-574. doi: 10.1038/s41423-024-01155-9. Epub 2024 Apr 3. Cell Mol Immunol. 2024. PMID: 38570588 - Experimental reconstitution of chronic ER stress in the liver reveals feedback suppression of BiP mRNA expression.
Gomez JA, Rutkowski DT. Gomez JA, et al. Elife. 2016 Dec 10;5:e20390. doi: 10.7554/eLife.20390. Elife. 2016. PMID: 27938665 Free PMC article.
References
- Ron D, Walter P. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous