Detection of Enhancer-Associated Rearrangements Reveals Mechanisms of Oncogene Dysregulation in B-cell Lymphoma - PubMed (original) (raw)
. 2015 Oct;5(10):1058-71.
doi: 10.1158/2159-8290.CD-15-0370. Epub 2015 Jul 30.
Yotam Drier 1, Holly Whitton 2, M Joel Cotton 1, Jasleen Kaur 1, Robbyn Issner 2, Shawn Gillespie 1, Charles B Epstein 2, Valentina Nardi 3, Aliyah R Sohani 3, Ephraim P Hochberg 4, Bradley E Bernstein 5
Affiliations
- PMID: 26229090
- PMCID: PMC4592453
- DOI: 10.1158/2159-8290.CD-15-0370
Detection of Enhancer-Associated Rearrangements Reveals Mechanisms of Oncogene Dysregulation in B-cell Lymphoma
Russell J H Ryan et al. Cancer Discov. 2015 Oct.
Abstract
B-cell lymphomas frequently contain genomic rearrangements that lead to oncogene activation by heterologous distal regulatory elements. We used a novel approach called "pinpointing enhancer-associated rearrangements by chromatin immunoprecipitation," or PEAR-ChIP, to simultaneously map enhancer activity and proximal rearrangements in lymphoma cell lines and patient biopsies. This method detects rearrangements involving known cancer genes, including CCND1, BCL2, MYC, PDCD1LG2, NOTCH1, CIITA, and SGK1, as well as novel enhancer duplication events of likely oncogenic significance. We identify lymphoma subtype-specific enhancers in the MYC locus that are silenced in lymphomas with MYC-activating rearrangements and are associated with germline polymorphisms that alter lymphoma risk. We show that BCL6-locus enhancers are acetylated by the BCL6-activating transcription factor MEF2B, and can undergo genomic duplication, or target the MYC promoter for activation in the context of a "pseudo-double-hit" t(3;8)(q27;q24) rearrangement linking the BCL6 and MYC loci. Our work provides novel insights regarding enhancer-driven oncogene activation in lymphoma.
Significance: We demonstrate a novel approach for simultaneous detection of genomic rearrangements and enhancer activity in tumor biopsies. We identify novel mechanisms of enhancer-driven regulation of the oncogenes MYC and BCL6, and show that the BCL6 locus can serve as an enhancer donor in an "enhancer hijacking" translocation.
©2015 American Association for Cancer Research.
Figures
Figure 1
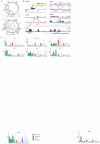
Detection of rearrangements involving the CCND1 and MYC loci by PEAR-ChIP. A. Left - Schematic depiction of a rearrangement between two chromosomes (red and green) with the breakpoint located in chromatin marked by H3K27ac (purple triangles). ChIP-Seq leads to isolation of H3K27ac-associated DNA, including enhancer-associated breakpoints. Right – Sequencing and alignment of ChIP DNA from case MCL-01 identifies read pairs at the ends of fragments containing the t(11;14) breakpoint (orange reads) and a physiological IGH VDJ recombination (red reads). B. Tracks showing H3K27ac signal in normal B cells (salmon), MCL cell lines (maroon) and MCL biopsies (dark purple) at the IGH and CCND1 loci. IGH-CCND1 rearrangement breakpoints detected by PEAR-ChIP are marked with black arrowheads. Also shown are breakpoints corresponding to intra-chromosomal deletions (paired open arrowheads) or large-scale rearrangements (single open arrowheads) affecting the CCND1 3’UTR. C. H3K27ac tracks and location of PEAR-ChIP-detected large-scale rearrangements (black arrows) involving the MYC locus.
Figure 2
Genome-wide rearrangement detection by PEAR-ChIP. A. Circos diagrams summarizing inter-chromosomal (dark shades) and intra-chromosomal (light shades) genomic rearrangements detected by H3K27ac PEAR-ChIP in 8 lymphoma cell lines (top) and 14 patient biopsies (bottom). Gene labels mark selected loci of interest. B. H3K27ac tracks and location of large-scale rearrangements (black arrows) detected by PEAR-ChIP at known oncogene (BCL2 and PDCD1LG2) and tumor suppressor gene (SGK1 and CIITA) loci, as well as a tandem duplication encompassing candidate enhancers upstream of TAGAP. Red bars link divergent read pairs spanning the tandem duplication. C. Normalized RNA expression (RQ = relative quantity) for genes affected by PEAR-ChIP-detected genomic lesions in HGB biopsies. Numbers at bottom denote the HGB sample name (e.g. “1” = HGB-01). Symbols indicate samples that contain a genomic lesion proximal to the measured gene (asterisk: inter-chromosomal rearrangement predicted to activate gene expression; minus sign: rearrangement predicted to inactivate gene expression; plus sign: enhancer tandem duplication). The pound sign denotes a FISH-detected rearrangement between the MYC and BCL6 loci. Samples are color-coded by gene expression subtype as follows: green = GCB-DLBCL, red = ABC-DLBCL, blue = PMBL, gray = unclassified. Solid bars denote samples studied by PEAR-ChIP.
Figure 3
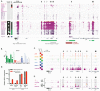
Acetylation and rearrangement of BCL6 locus enhancers. A. H3K27ac ChIP-Seq tracks across the BCL6 locus in 29 B cell populations. Green bars at bottom indicate the median positions of detected super-enhancer regions. Black brackets indicate published breakpoint cluster regions. Read-pairs spanning the tandem duplication in HGB-04 are linked by red bars at bottom. The legend at left indicates the super-enhancers called in each population, as well as populations classified as HGB (red = ABC-DLBCL, green = GCB-DLBCL, blue = PMBL, black = HGB cell line). MEF2B-positive (black arrows) and selected MEF2B-negative TF binding sites (open arrows) are indicated, and correspond to positions indicated in (C). ChIP-seq coverage range is 0-5 fpm (fragments per million mapped fragments) for all tracks. B. Normalized RNA expression of BCL6 in HGB biopsies. Samples are numbered and color-coded by subtype as in Figure 2C. Solid boxes indicate samples evaluated by PEAR-ChIP. Symbols indicate samples that contain a genomic lesion proximal to the measured gene (plus sign: enhancer tandem duplication, pound sign: FISH-detected rearrangement between the MYC and BCL6 loci). C. ChIP-Seq tracks for p300, H3K27ac, and three TFs in two HGB cell lines at the BCL6 promoter and super-enhancer regions. MEF2B-positive (black arrows) and selected MEF2B-negative TF binding sites (open arrows) are indicated and correspond to positions indicated in (A). ChIP-Seq coverage range is 0-15 fpm for H3K27ac. D. Relative BCL6 expression in MCL and HGB cell lines stably transduced with pINDUCER-20 bearing a GFP or MEF2B-HA transgene, and harvested 48 hours after induction with 100 ng/ml doxycycline. E. ChIP-seq tracks for H3K27ac and HA-tag in Jeko-1 cells with induced expression of GFP or MEF2B-HA as in figure 4b. Genomic positions and arrows are identical to (C). ChIP-seq coverage range is 0-2.5 fpm for H3K27ac.
Figure 4
Enhancer acetylation at native and rearranged MYC loci in B-cell lymphoma. A. Interaction of candidate MYC locus enhancer regions with the MYC promoter by chromatin conformation capture (3C) in HGB cell lines (top), and corresponding H3K27ac ChIP-Seq tracks (bottom). Red bar at bottom indicates 3’ _MYC_-interacting region detailed in (C). Arrows indicate the positions of previously reported MYC enhancers in T cell leukemia (black arrow) (30) and myeloid leukemia (open arrow) (3). ChIP-seq coverage range is 0-10 fpm. B. Interaction of candidate MYC locus enhancer regions with the MYC promoter by chromatin conformation capture (3C) in MCL and SLL biopsies (top), and corresponding H3K27ac ChIP-Seq tracks (bottom). Red bar at bottom indicates 5’ _MYC_-interacting region detailed in (C). ChIP-seq coverage range is 0-5 fpm. C. Detail of H3K27ac ChIP-seq tracks at _MYC_-interacting regions in B cell samples. Legend at center indicates lymphoma type (for HGB biopsies red = ABC-DLBCL, green = GCB-DLBCL, blue = PMBL) or normal B population, presence or absence of a MYC rearrangement by fluorescence in situ hybridization (“MYC FISH”, orange: rearrangement, gray: not detected, white: not evaluated), and MYC rearrangement detection by PEAR-ChIP (“PC MYC-R”). Triangles at bottom indicate position and lymphoma subtype associations of single-nucleotide polymorphisms linked to hereditary risk for lymphoma in published GWAS studies. ChIP-seq coverage range is 0-5 fpm for all tracks.
Figure 5
A t(3;8)(q27;q24) rearrangement leads to MYC activation by BCL6 enhancers. A. H3K27ac ChIP-Seq tracks at the BCL6 locus for HGB-06 and HGB-07. The MYC promoter region is also shown for HGB-07, and a dashed black line connects the t(3;8) rearrangement breakpoints. Solid black (HGB-06) or red (HGB-07) arrows depict looping interactions on the native chromosome 3 as determined by 3C. Dashed red arrows depict looping interactions between BCL6 enhancers and the MYC promoter in HGB-07, as determined by 3C. B. H3K27ac ChIP-Seq tracks at the MYC locus for HGB-06 and HGB-07. The BCL6 promoter region is also shown for HGB-07, and a dashed black line connects the t(3;8) rearrangement breakpoints in HGB-07. Solid black (HGB-06) or red (HGB-07) arrows depict looping interactions on the native chromosome 8 as determined by 3C. C. 3C data for interactions between BCL6 locus enhancers and the BCL6 promoter (top) or MYC promoter (bottom) for HGB-06 and HGB-07. “B” indicates the position of the breakpoint junction on the der(8)t(3;8) in HGB-07. D. 3C data for interactions between MYC locus distal elements and the MYC promoter (top) or BCL6 promoter (bottom) for HGB-06 and HGB-07. “B” indicates the position of the breakpoint junction on the der(3)t(3;8) in HGB-07. E. Schematic models of MYC promoter interactions with native and heterologous enhancers in B cell lymphomas.
Comment in
- "PEAR-ing" Genomic and Epigenomic Analyses for Cancer Gene Discovery.
Mack SC, Rich JN, Scacheri PC. Mack SC, et al. Cancer Discov. 2015 Oct;5(10):1018-20. doi: 10.1158/2159-8290.CD-15-0985. Cancer Discov. 2015. PMID: 26429935 Free PMC article.
Similar articles
- A novel five-way translocation, t(3;9;13;8;14)(q27;p13;q32;q24;q32), with concurrent MYC and BCL6 rearrangements in a primary bone marrow B-cell lymphoma.
Yamamoto K, Matsuoka H, Yakushijin K, Funakoshi Y, Okamura A, Hayashi Y, Minami H. Yamamoto K, et al. Cancer Genet. 2011 Sep;204(9):501-6. doi: 10.1016/j.cancergen.2011.08.017. Cancer Genet. 2011. PMID: 22018272 - Clinical relevance of BCL2, BCL6, and MYC rearrangements in diffuse large B-cell lymphoma.
Kramer MH, Hermans J, Wijburg E, Philippo K, Geelen E, van Krieken JH, de Jong D, Maartense E, Schuuring E, Kluin PM. Kramer MH, et al. Blood. 1998 Nov 1;92(9):3152-62. Blood. 1998. PMID: 9787151 - Translocation (3;8)(q27;q24) in two cases of triple hit lymphoma.
Motlló C, Grau J, Juncà J, Ruiz N, Mate JL, Orna E, Navarro JT, Vives S, Sancho JM, Esteban D, Granada I, Feliu E, Ribera JM, Millá F. Motlló C, et al. Cancer Genet Cytogenet. 2010 Dec;203(2):328-32. doi: 10.1016/j.cancergencyto.2010.08.018. Cancer Genet Cytogenet. 2010. PMID: 21156254 - The BCL6 proto-oncogene: a leading role during germinal center development and lymphomagenesis.
Jardin F, Ruminy P, Bastard C, Tilly H. Jardin F, et al. Pathol Biol (Paris). 2007 Feb;55(1):73-83. doi: 10.1016/j.patbio.2006.04.001. Epub 2006 Jul 3. Pathol Biol (Paris). 2007. PMID: 16815642 Review. - MYC-associated and double-hit lymphomas: a review of pathobiology, prognosis, and therapeutic approaches.
Petrich AM, Nabhan C, Smith SM. Petrich AM, et al. Cancer. 2014 Dec 15;120(24):3884-95. doi: 10.1002/cncr.28899. Epub 2014 Jul 24. Cancer. 2014. PMID: 25060588 Review.
Cited by
- Mutant p53 regulates enhancer-associated H3K4 monomethylation through interactions with the methyltransferase MLL4.
Rahnamoun H, Hong J, Sun Z, Lee J, Lu H, Lauberth SM. Rahnamoun H, et al. J Biol Chem. 2018 Aug 24;293(34):13234-13246. doi: 10.1074/jbc.RA118.003387. Epub 2018 Jun 28. J Biol Chem. 2018. PMID: 29954944 Free PMC article. - BRD4 and MYC: power couple in transcription and disease.
Kotekar A, Singh AK, Devaiah BN. Kotekar A, et al. FEBS J. 2023 Oct;290(20):4820-4842. doi: 10.1111/febs.16580. Epub 2022 Aug 3. FEBS J. 2023. PMID: 35866356 Free PMC article. Review. - The 3D enhancer network of the developing T cell genome is shaped by SATB1.
Zelenka T, Klonizakis A, Tsoukatou D, Papamatheakis DA, Franzenburg S, Tzerpos P, Tzonevrakis IR, Papadogkonas G, Kapsetaki M, Nikolaou C, Plewczynski D, Spilianakis C. Zelenka T, et al. Nat Commun. 2022 Nov 14;13(1):6954. doi: 10.1038/s41467-022-34345-y. Nat Commun. 2022. PMID: 36376298 Free PMC article. - Characteristics and Clinical Value of MYC , BCL2, and BCL6 Rearrangement Detected by Next-generation Sequencing in DLBCL.
Zeng Y, Wei R, Bao L, Xue T, Qin Y, Ren M, Bai Q, Yao Q, Yu C, Chen C, Wei P, Yu B, Cao J, Li X, Zhang Q, Zhou X. Zeng Y, et al. Am J Surg Pathol. 2024 Aug 1;48(8):919-929. doi: 10.1097/PAS.0000000000002258. Epub 2024 Jun 28. Am J Surg Pathol. 2024. PMID: 38937822 Free PMC article. - Inherited variants at 3q13.33 and 3p24.1 are associated with risk of diffuse large B-cell lymphoma and implicate immune pathways.
Kleinstern G, Yan H, Hildebrandt MAT, Vijai J, Berndt SI, Ghesquières H, McKay J, Wang SS, Nieters A, Ye Y, Monnereau A, Brooks-Wilson AR, Lan Q, Melbye M, Jackson RD, Teras LR, Purdue MP, Vajdic CM, Vermeulen RCH, Giles GG, Cocco PL, Birmann BM, Kraft P, Albanes D, Zeleniuch-Jacquotte A, Crouch S, Zhang Y, Sarangi V, Asmann Y, Offit K, Salles G, Wu X, Smedby KE, Skibola CF, Slager SL, Rothman N, Chanock SJ, Cerhan JR. Kleinstern G, et al. Hum Mol Genet. 2020 Jan 1;29(1):70-79. doi: 10.1093/hmg/ddz228. Hum Mol Genet. 2020. PMID: 31600786 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- T32CA009216/CA/NCI NIH HHS/United States
- T32 CA009216/CA/NCI NIH HHS/United States
- U54 HG004570/HG/NHGRI NIH HHS/United States
- T32GM007748/GM/NIGMS NIH HHS/United States
- T32 GM007748/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials