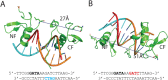GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA - PubMed (original) (raw)
. 2015 Oct;24(10):1649-59.
doi: 10.1002/pro.2760. Epub 2015 Aug 20.
Affiliations
- PMID: 26234528
- PMCID: PMC4594664
- DOI: 10.1002/pro.2760
GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA
Lorna Wilkinson-White et al. Protein Sci. 2015 Oct.
Abstract
The transcription factor GATA1 helps regulate the expression of thousands of genes involved in blood development, by binding to single or double GATA sites on DNA. An important part of gene activation is chromatin looping, the bringing together of DNA elements that lie up to many thousands of basepairs apart in the genome. It was recently suggested, based on studies of the closely related protein GATA3, that GATA-mediated looping may involve interactions of each of two zinc fingers (ZF) with distantly spaced DNA elements. Here we present a structure of the GATA1 ZF region bound to pseudopalindromic double GATA site DNA, which is structurally equivalent to a recently-solved GATA3-DNA complex. However, extensive analysis of GATA1-DNA binding indicates that although the N-terminal ZF (NF) can modulate GATA1-DNA binding, under physiological conditions the NF binds DNA so poorly that it cannot play a direct role in DNA-looping. Rather, the ability of the NF to stabilize transcriptional complexes through protein-protein interactions, and thereby recruit looping factors such as Ldb1, provides a more compelling model for GATA-mediated looping.
Keywords: DNA binding; GATA1; chromatin looping; protein-DNA structure; transcription factor complex.
© 2015 The Protein Society.
Figures
Figure 1
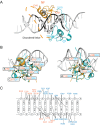
GATA1NC binds pseudopalindromic DNA in a 1:1 complex. (A) Overall structure of the GATA1NC:mPal Complex (3vd6). Ribbon diagram of GATA1NC bound to an mPal containing oligonucleotide (white and black to indicate the flanking and core mPAL sequence, respectively; the nucleotides in the core sequence are labelled). The NF is shown in orange and the CF is shown in cyan, with the zinc atoms and zinc-ligating sidechains shown as grey spheres and sticks. The linker region between the two fingers was not present in the electron density map and is represented by an orange dashed line. (B) The GATA1NF (orange) aligned with the GATA1CF (cyan) bound to DNA (white) shown from two angles. All residues that are identical in the sequences of the two domains are shown as sticks; residues discussed in the main text are labelled. (C) Schematic of DNA-protein interactions. Orange labels indicate NF and blue labels indicate CF interactions. Bold lines—hydrogen bonds between amino acids and bases; dashed lines—hydrophobic interactions; Dotted lines—contacts with sugar-phosphate backbone. An interactive view is available in the electronic version of the article.
Figure 2
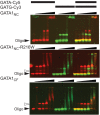
GATA1 is unable to bridge binding sites on separate DNA strands. EMSA analysis of GATA1NC, GATA1NC-R216W and GATA1CF binding to GATA-Cy5 (protein concentrations of 50, 100, 200, 400, and 600 n_M_ protein, top and bottom panels also include 20 n_M_ protein) and GATG-Cy3 (50, 100, 200, 400, and 600 n_M_ protein) and both the GATA-Cy5 and GATG-Cy3 simultaneously (20, 50, 100, 200, 400, and 600 n_M_ protein). Each oligonucleotide was at 20 n_M_. Images show the overlay of Cy5 (red) and Cy3 (green) fluorescence. The solid triangle indicates unbound DNA, the open triangle indicates GATA1CF bound to DNA. Other complexes are discussed in the text.
Figure 3
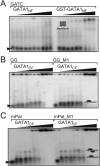
GATA1CF but not GATA1NF shows robust binding to DNA. EMSA analysis of (A) GATA1NF (lanes 2–10) or GST-GATA1NF (lanes 13–20) binding to a GATC-containing oligonucleotide at concentrations of 100, 200, 400, 600, 800, 1000, 2000, and 3000 n_M_ at low salt (15 m_M_ NaCl); and GATA1CF binding to (B) GG (lanes 2–10) or GG_M1 (lanes 12–20) at concentrations of 50, 100, 200, 400, 500, 800, 1000, 2000, 5000, and 7500 n_M_ in 15 m_M_ NaCl, or (C) to mPal (lanes 2–10) or mPal_M1 (lanes 12–20) at concentrations of 50, 100, 200, 400, 500, 800, 1000, 2000, 5000, and 7500 n_M_ in 15 m_M_ NaCl.
Figure 4
EMSA analysis of GATA1NC binding to double-site DNA. Binding of GATA1NC to (A) mPal (lanes 2–10) or mPal _M1 (lanes 12–20) at 10, 20, 50, 100, 200, 400, 600, 800, and 1000 n_M_ on low salt (15 m_M_ NaCl), (B) mPal (lanes 2–8) or mPal_M1 (lanes 10–15) at concentrations of 0.5, 1, 2, 3, 4, 5, and 6 µ_M_ in physiological salt (150 m_M_ NaCl) (C) GG at 20, 50, 100, 200, 400, and 600 n_M_ (lanes 2–7), GG_M1 oligonucleotide at 100, 200, 400, 600, 800, and 1000 n_M_ (lanes 9–14) and to a single site GATA oligonucleotide (1GATA) at 50, 100, 200, 400, and 600 n_M_ (lanes 16–20) in low salt (15 m_M_ NaCl), (D) GG (lanes 2–8) and GG_M1 (lanes 9–16) at 0.5, 1, 2, 3, 4, 5, and 6 µ_M_ in physiological salt (150 m_M_ NaCl). Schematics of complexes (indicated with numbered arrows) show the GATA1NF in white and GATA1CF in black. Note that the change in intensity throughout the final lane of panel C is an artefact from phosphorimaging.
Figure 5
Model of GATA1NC in complex with GG DNA. (A) Model in which the NF contacts the GATC site (cyan) on the 3′ → 5′ strand. (B) Model in which the NF contacting the GATC site (red) on the 5′ → 3′ strand. The DNA sequence shown with the binding motif indicated. The minimum distances between the ends of the NF and CF are indicated.
Similar articles
- Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor.
Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Deng W, et al. Cell. 2012 Jun 8;149(6):1233-44. doi: 10.1016/j.cell.2012.03.051. Cell. 2012. PMID: 22682246 Free PMC article. - GATA zinc finger interactions modulate DNA binding and transactivation.
Trainor CD, Ghirlando R, Simpson MA. Trainor CD, et al. J Biol Chem. 2000 Sep 8;275(36):28157-66. doi: 10.1074/jbc.M000020200. J Biol Chem. 2000. PMID: 10862757 - GATA1 Binding Kinetics on Conformation-Specific Binding Sites Elicit Differential Transcriptional Regulation.
Hasegawa A, Kaneko H, Ishihara D, Nakamura M, Watanabe A, Yamamoto M, Trainor CD, Shimizu R. Hasegawa A, et al. Mol Cell Biol. 2016 Jul 29;36(16):2151-67. doi: 10.1128/MCB.00017-16. Print 2016 Aug 15. Mol Cell Biol. 2016. PMID: 27215385 Free PMC article. - Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1.
Wilkinson-White L, Gamsjaeger R, Dastmalchi S, Wienert B, Stokes PH, Crossley M, Mackay JP, Matthews JM. Wilkinson-White L, et al. Proc Natl Acad Sci U S A. 2011 Aug 30;108(35):14443-8. doi: 10.1073/pnas.1105898108. Epub 2011 Aug 15. Proc Natl Acad Sci U S A. 2011. PMID: 21844373 Free PMC article. - GATA-1: friends, brothers, and coworkers.
Morceau F, Schnekenburger M, Dicato M, Diederich M. Morceau F, et al. Ann N Y Acad Sci. 2004 Dec;1030:537-54. doi: 10.1196/annals.1329.064. Ann N Y Acad Sci. 2004. PMID: 15659837 Review.
Cited by
- A Distinct Mechanism of RNA Recognition by the Transcription Factor GATA1.
Ugay DA, Batey RT, Wuttke DS. Ugay DA, et al. Biochemistry. 2025 Mar 18;64(6):1193-1198. doi: 10.1021/acs.biochem.4c00818. Epub 2025 Feb 25. Biochemistry. 2025. PMID: 39999571 - SENP1 promotes triple-negative breast cancer invasion and metastasis via enhancing CSN5 transcription mediated by GATA1 deSUMOylation.
Gao Y, Wang R, Liu J, Zhao K, Qian X, He X, Liu H. Gao Y, et al. Int J Biol Sci. 2022 Mar 6;18(5):2186-2201. doi: 10.7150/ijbs.60594. eCollection 2022. Int J Biol Sci. 2022. PMID: 35342335 Free PMC article. - Disparate binding kinetics by an intrinsically disordered domain enables temporal regulation of transcriptional complex formation.
Robertson NO, Smith NC, Manakas A, Mahjoub M, McDonald G, Kwan AH, Matthews JM. Robertson NO, et al. Proc Natl Acad Sci U S A. 2018 May 1;115(18):4643-4648. doi: 10.1073/pnas.1714646115. Epub 2018 Apr 16. Proc Natl Acad Sci U S A. 2018. PMID: 29666277 Free PMC article. - GATA1 Promotes Gemcitabine Resistance in Pancreatic Cancer through Antiapoptotic Pathway.
Chang Z, Zhang Y, Liu J, Guan C, Gu X, Yang Z, Ye Q, Ding L, Liu R. Chang Z, et al. J Oncol. 2019 Apr 10;2019:9474273. doi: 10.1155/2019/9474273. eCollection 2019. J Oncol. 2019. PMID: 31093285 Free PMC article. - GATA factor mutations in hematologic disease.
Crispino JD, Horwitz MS. Crispino JD, et al. Blood. 2017 Apr 13;129(15):2103-2110. doi: 10.1182/blood-2016-09-687889. Epub 2017 Feb 8. Blood. 2017. PMID: 28179280 Free PMC article. Review.
References
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the [beta]-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. - PubMed
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources