Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging - PubMed (original) (raw)
Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging
Tanmay A M Bharat et al. Structure. 2015.
Abstract
Recent innovations in specimen preparation, data collection, and image processing have led to improved structure determination using single-particle electron cryomicroscopy (cryo-EM). Here we explore some of these advances to improve structures determined using electron cryotomography (cryo-ET) and sub-tomogram averaging. We implement a new three-dimensional model for the contrast transfer function, and use this in a regularized likelihood optimization algorithm as implemented in the RELION program. Using direct electron detector data, we apply both single-particle analysis and sub-tomogram averaging to analyze radiation-induced movements of the specimen. As in single-particle cryo-EM, we find that significant sample movements occur during tomographic data acquisition, and that these movements are substantially reduced through the use of ultrastable gold substrates. We obtain a sub-nanometer resolution structure of the hepatitis B capsid, and show that reducing radiation-induced specimen movement may be central to attempts at further improving tomogram quality and resolution.
Copyright © 2015 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
Figure 1
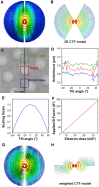
3D CTF Model and CTF Estimation (A) An isosurface view of the 3D CTF model used. The volume has been pseudo-colored based on radius (or resolution in Fourier space). Red indicates low resolution while blue indicates high resolution. The 3D CTF model is made of a series of 2D slices that represent 2D CTFs of each image of the tilt series. (B) An orthogonal view of (A) with only the central slice shown. (C) A low-magnification micrograph showing the extended tilt series acquisition used in this study. Two additional images were acquired to estimate the CTF parameters in the target region of interest. (D) A plot of estimated defoci in each of the three regions shown in (C) at different tilt angles. The difference between the green and the blue curves might be caused by an inclination of the sample with respect to the tilt axis. The diameter of holes in the micrograph is 2 µm. (E) Tilt angle-dependent scaling factor applied to weight the 3D CTF model. The multiplicative factor is equal to the cosine of the tilt angle, and scales the entire CTF curve downward. (F) Dose-dependent _B_–factor applied to the CTF model. The slope of the linear curve was determined empirically from a previous single-particle analysis report (Scheres, 2014). (G and H) Isosurface view (G) and central slice (H) of the final weighted 3D CTF model.
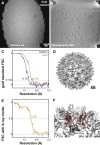
Figure 2
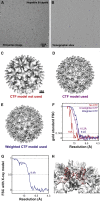
Maximum-Likelihood Refinement Using the 3D CTF Model (A) Representative image from a tilt series of the hepatitis B capsid sample. (B) Slice from a tomogram reconstructed from the tilt series data. The scale of (A) and (B) is the same. (C–E) Isosurface representations of the final volume of the refinement calculated (C) without CTF correction, (D) with the unweighted CTF model, and (E) with the weighted CTF model. The scale bar in (C) also applies to (D) and (E). (F) FSC curves showing the estimated resolution from the refinements. The same 1,851 HBV particles from 15 tomograms were used in all refinements. (G) FSC curve of the best model (from the refinement with weighted CTF model) against the X-ray structure. (H) The X-ray structure is shown fitted into the sub-tomogram averaging map, and secondary structure elements are clearly resolved. See also Figure S1.
Figure 3
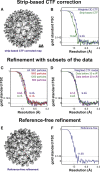
Refinements with Differing Parameters to Test the 3D CTF Model and to Probe the Limits of the Dataset (A) Using conventional strip-based CTF correction implemented in IMOD, automatic refinement in RELION yielded a 10.9 Å resolution map. Only phase flipping and no amplitude weighting was conducted in this CTF correction. (B) Compared with the refinement using our new 3D CTF model, the resolution obtained was lower. (C) Resolution of sub-tomogram averaging reconstruction could not be improved by adding more data or removing subsets of the data. Different sub-tomogram averaging refinements were conducted using random subsets of the data of different sizes. A similar resolution was obtained with fewer particles, showing that the refinement was not limited by the size of the dataset. (D) Part of the data that had been exposed to a cumulative electron dose of > 20 or 30 e−/Å2 was removed using the 3D CTF model. Compared with the full dataset in which 60 e−/Å2 had been applied to the specimen, no improvement was observed (the weighted 3D CTF model was used in all refinements). (E) Reference-free refinement using the same data as shown in Figure 2. Scale same as (A). (F) The resolution of the output, refined structure (11.5 Å) was not as high as in cases when a reference was used (9.2 Å).
Figure 4
Pre-illumination for Comparing Sub-tomogram Averaging and Single-Particle Analysis (A) The sample was pre-illuminated with a dose of 6 e−/Å2 to record an image for single-particle analysis (SPA). (B) Immediately after this, a tilt series was collected of the same region with the same microscope parameters. The scale bar applies to (A) and (B). (C) Sub-tomogram averaging (STA) was conducted from the tomographic data, and single-particle analysis was conducted from the pre-illumination image using the same 1,501 HBV particles from 13 tomograms in both refinements. FSC curves of these refinements are shown. (D) Sub-tomogram averaging refinement of half of the particles (n = 751) that had the most similar alignment parameters in both refinements (denoted as subset 1, also see Figure S2) were compared with the other half (subset 2, n = 750) that had the most different alignment parameters. There was no significant difference between the resolutions obtained in both refinements. Furthermore, applying Euler angles obtained from single-particle analysis to the entire dataset (of 1,501 particles) did not lead to an improvement in sub-tomogram averaging (purple curve). See also Figure S2.
Figure 5
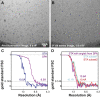
Sub-tomogram Averaging with Reduced Radiation-Induced Motion (A) Representative image from a tilt series of the hepatitis B capsid sample imaged on a gold support grid. The support is seen as a high-contrast feature. (B) Slice from a tomogram reconstructed from the tilt series data. The scale bar applies to (A) and (B). (C) FSC curves showing the estimated resolution from the refinements of data from carbon grids and gold supports. (1,145 HBV capsid particles were used in both refinements; for the gold supports these came from 11 tomograms.) (D) Isosurface representation of the output volume from the refinement with data collected on gold supports. (E) FSC curve of the sub-tomogram averaging model from the refinement with data collected on gold supports against the X-ray structure. (F) The X-ray structure is shown fitted into the sub-tomogram averaging map where secondary structure elements are clearly resolved. See also Figures S3 and S4.
Similar articles
- Towards high-throughput in situ structural biology using electron cryotomography.
Böhning J, Bharat TAM. Böhning J, et al. Prog Biophys Mol Biol. 2021 Mar;160:97-103. doi: 10.1016/j.pbiomolbio.2020.05.010. Epub 2020 Jun 21. Prog Biophys Mol Biol. 2021. PMID: 32579969 Review. - Processing of Structurally Heterogeneous Cryo-EM Data in RELION.
Scheres SH. Scheres SH. Methods Enzymol. 2016;579:125-57. doi: 10.1016/bs.mie.2016.04.012. Epub 2016 May 31. Methods Enzymol. 2016. PMID: 27572726 Review. - Rapid increase of near atomic resolution virus capsid structures determined by cryo-electron microscopy.
Ho PT, Reddy VS. Ho PT, et al. J Struct Biol. 2018 Jan;201(1):1-4. doi: 10.1016/j.jsb.2017.10.007. Epub 2017 Oct 27. J Struct Biol. 2018. PMID: 29080674 - Alignment of cryo-EM movies of individual particles by optimization of image translations.
Rubinstein JL, Brubaker MA. Rubinstein JL, et al. J Struct Biol. 2015 Nov;192(2):188-95. doi: 10.1016/j.jsb.2015.08.007. Epub 2015 Aug 19. J Struct Biol. 2015. PMID: 26296328 - Subtomogram averaging from cryo-electron tomograms.
Leigh KE, Navarro PP, Scaramuzza S, Chen W, Zhang Y, Castaño-Díez D, Kudryashev M. Leigh KE, et al. Methods Cell Biol. 2019;152:217-259. doi: 10.1016/bs.mcb.2019.04.003. Epub 2019 May 15. Methods Cell Biol. 2019. PMID: 31326022 Review.
Cited by
- CryoDRGN-ET: deep reconstructing generative networks for visualizing dynamic biomolecules inside cells.
Rangan R, Feathers R, Khavnekar S, Lerer A, Johnston JD, Kelley R, Obr M, Kotecha A, Zhong ED. Rangan R, et al. Nat Methods. 2024 Aug;21(8):1537-1545. doi: 10.1038/s41592-024-02340-4. Epub 2024 Jul 18. Nat Methods. 2024. PMID: 39025970 - Complete atomic structure of a native archaeal cell surface.
von Kügelgen A, Alva V, Bharat TAM. von Kügelgen A, et al. Cell Rep. 2021 Nov 23;37(8):110052. doi: 10.1016/j.celrep.2021.110052. Cell Rep. 2021. PMID: 34818541 Free PMC article. - High-resolution in situ structure determination by cryo-electron tomography and subtomogram averaging using emClarity.
Ni T, Frosio T, Mendonça L, Sheng Y, Clare D, Himes BA, Zhang P. Ni T, et al. Nat Protoc. 2022 Feb;17(2):421-444. doi: 10.1038/s41596-021-00648-5. Epub 2022 Jan 12. Nat Protoc. 2022. PMID: 35022621 Free PMC article. Review. - In situ cryo-ET structure of phycobilisome-photosystem II supercomplex from red alga.
Li M, Ma J, Li X, Sui SF. Li M, et al. Elife. 2021 Sep 13;10:e69635. doi: 10.7554/eLife.69635. Elife. 2021. PMID: 34515634 Free PMC article. - Learning structural heterogeneity from cryo-electron sub-tomograms with tomoDRGN.
Powell BM, Davis JH. Powell BM, et al. bioRxiv [Preprint]. 2023 Jun 2:2023.05.31.542975. doi: 10.1101/2023.05.31.542975. bioRxiv. 2023. PMID: 37398315 Free PMC article. Updated. Preprint.
References
- Agulleiro J.I., Fernandez J.J. Fast tomographic reconstruction on multicore computers. Bioinformatics. 2011;27:582–583. - PubMed
- Bai X.C., McMullan G., Scheres S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015;40:49–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U105192715/MRC_/Medical Research Council/United Kingdom
- MC_UP_A025_1013/MRC_/Medical Research Council/United Kingdom
- MC_U105184326/MRC_/Medical Research Council/United Kingdom
- MC_UPA0251013/MRC_/Medical Research Council/United Kingdom
- 261151/ERC_/European Research Council/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials