Replicating Nucleosomes - PubMed (original) (raw)
Replicating Nucleosomes
Srinivas Ramachandran et al. Sci Adv. 2015.
Abstract
Eukaryotic replication disrupts each nucleosome as the fork passes, followed by re-assembly of disrupted nucleosomes and incorporation of newly synthesized histones into nucleosomes in the daughter genomes. In this review, we examine this process of replication-coupled nucleosome assembly to understand how characteristic steady state nucleosome landscapes are attained. Recent studies have begun to elucidate mechanisms involved in histone transfer during replication and maturation of the nucleosome landscape after disruption by replication. A fuller understanding of replication-coupled nucleosome assembly will be needed to explain how epigenetic information is replicated at every cell division.
Keywords: epigenetics; histone modification; histone variants; replication-coupled nucleosome assembly.
Figures
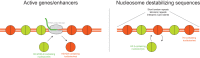
Fig. 1. Distinct mechanisms of incorporation of H3.3 at active and silent regions of the genome.
At transcriptionally active regions, nucleosomes are disrupted by RNA polymerase or nucleosome remodelers, resulting in occasional nucleosome loss, with replacement by nucleosomes containing H3.3, which unlike H3 is present throughout the cell cycle. At regions of unusual base composition, which include telomeres, CpG islands, and short-period satellite repeats, the lack of nucleosome-stabilizing sequences results in relatively frequent nucleosome loss, with replacement by H3.3 nucleosomes.
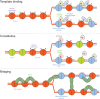
Fig. 2. Three models of propagating histone modifications through replication.
In the template-binding model (42), adjacent nucleosomes are modified by a histone-modifying enzyme that binds the modified residue on a nearby tail. In the constitutive model (47), H3K27 methylation is restored by recognition of H3A31 but not H3.3T31 by ATXR5/6, such that only replication-coupled (H3) nucleosomes, not replication-independent (H3.3) nucleosomes, are methylated on H3K27. In the bridging model (48), PRC1 bridges nucleosomes across daughter chromatids.
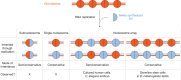
Fig. 3. Different modes of inheritance of old H3 and newly synthesized H3 on newly replicated DNA.
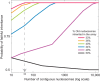
Fig. 4. Probability of faithful inheritance at various thresholds of old histone segregation.
Given that old (H3-H4)2 is segregated randomly to daughter chromosomes during replication, we can think of each nucleosome assembled as a Bernoulli trial, with the probability of a daughter chromosome assembling a nucleosome with old (H3-H4)2 the same as the probability of assembling a nucleosome with new (H3-H4)2, both of which would be 0.5. We can then ask, what is the probability that at least a given percentage of old (H3-H4)2 in a nucleosome array of a given size is obtained by a daughter chromosome? The x axis of this plot represents the different nucleosome array sizes. The y axis represents the probability of the daughter chromosome getting at least a given percentage of old (H3-H4)2. We define this binomial probability as the probability of faithful inheritance. The dashed gray line represents the minimum size of the nucleosome array that would ensure faithful inheritance of at least 33% of old (H3-H4)2.
Similar articles
- The Role of Histone Modification in DNA Replication-Coupled Nucleosome Assembly and Cancer.
Zhang Y, Zhang Q, Zhang Y, Han J. Zhang Y, et al. Int J Mol Sci. 2023 Mar 3;24(5):4939. doi: 10.3390/ijms24054939. Int J Mol Sci. 2023. PMID: 36902370 Free PMC article. Review. - Histone-modifying enzymes, histone modifications and histone chaperones in nucleosome assembly: Lessons learned from Rtt109 histone acetyltransferases.
Dahlin JL, Chen X, Walters MA, Zhang Z. Dahlin JL, et al. Crit Rev Biochem Mol Biol. 2015 Jan-Feb;50(1):31-53. doi: 10.3109/10409238.2014.978975. Epub 2014 Nov 3. Crit Rev Biochem Mol Biol. 2015. PMID: 25365782 Free PMC article. Review. - Replication-Coupled Nucleosome Assembly in the Passage of Epigenetic Information and Cell Identity.
Serra-Cardona A, Zhang Z. Serra-Cardona A, et al. Trends Biochem Sci. 2018 Feb;43(2):136-148. doi: 10.1016/j.tibs.2017.12.003. Epub 2017 Dec 29. Trends Biochem Sci. 2018. PMID: 29292063 Free PMC article. Review. - Histones, histone chaperones and nucleosome assembly.
Burgess RJ, Zhang Z. Burgess RJ, et al. Protein Cell. 2010 Jul;1(7):607-12. doi: 10.1007/s13238-010-0086-y. Protein Cell. 2010. PMID: 21203931 Free PMC article. Review. - Single-molecule imaging reveals control of parental histone recycling by free histones during DNA replication.
Gruszka DT, Xie S, Kimura H, Yardimci H. Gruszka DT, et al. Sci Adv. 2020 Sep 18;6(38):eabc0330. doi: 10.1126/sciadv.abc0330. Print 2020 Sep. Sci Adv. 2020. PMID: 32948589 Free PMC article.
Cited by
- Histone variant H2A.Z regulates nucleosome unwrapping and CTCF binding in mouse ES cells.
Wen Z, Zhang L, Ruan H, Li G. Wen Z, et al. Nucleic Acids Res. 2020 Jun 19;48(11):5939-5952. doi: 10.1093/nar/gkaa360. Nucleic Acids Res. 2020. PMID: 32392318 Free PMC article. - Stable inheritance of H3.3-containing nucleosomes during mitotic cell divisions.
Xu X, Duan S, Hua X, Li Z, He R, Zhang Z. Xu X, et al. Nat Commun. 2022 May 6;13(1):2514. doi: 10.1038/s41467-022-30298-4. Nat Commun. 2022. PMID: 35523900 Free PMC article. - Repression of CENP-A assembly in metaphase requires HJURP phosphorylation and inhibition by M18BP1.
Flores Servin JC, Brown RR, Straight AF. Flores Servin JC, et al. J Cell Biol. 2023 Jun 5;222(6):e202110124. doi: 10.1083/jcb.202110124. Epub 2023 May 4. J Cell Biol. 2023. PMID: 37141119 Free PMC article. - From correlation to causation: The new frontier of transgenerational epigenetic inheritance.
Rothi MH, Greer EL. Rothi MH, et al. Bioessays. 2023 Jan;45(1):e2200118. doi: 10.1002/bies.202200118. Epub 2022 Nov 9. Bioessays. 2023. PMID: 36351255 Free PMC article. - Mechanisms of Nucleosome Dynamics In Vivo.
Henikoff S. Henikoff S. Cold Spring Harb Perspect Med. 2016 Sep 1;6(9):a026666. doi: 10.1101/cshperspect.a026666. Cold Spring Harb Perspect Med. 2016. PMID: 27503998 Free PMC article. Review.
References
- Kim T. K., Hemberg M., Gray J. M., Costa A. M., Bear D. M., Wu J., Harmin D. A., Laptewicz M., Barbara-Haley K., Kuersten S., Markenscoff-Papadimitriou E., Kuhl D., Bito H., Worley P. F., Kreiman G., Greenberg M. E., Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010). - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources