CTFFIND4: Fast and accurate defocus estimation from electron micrographs - PubMed (original) (raw)
CTFFIND4: Fast and accurate defocus estimation from electron micrographs
Alexis Rohou et al. J Struct Biol. 2015 Nov.
Abstract
CTFFIND is a widely-used program for the estimation of objective lens defocus parameters from transmission electron micrographs. Defocus parameters are estimated by fitting a model of the microscope's contrast transfer function (CTF) to an image's amplitude spectrum. Here we describe modifications to the algorithm which make it significantly faster and more suitable for use with images collected using modern technologies such as dose fractionation and phase plates. We show that this new version preserves the accuracy of the original algorithm while allowing for higher throughput. We also describe a measure of the quality of the fit as a function of spatial frequency and suggest this can be used to define the highest resolution at which CTF oscillations were successfully modeled.
Keywords: Astigmatism; CTF; Defocus; Phase plate.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
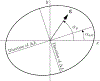
Figure 1:
Two defocus values, Δf1 and Δf2, and an angle, α ast define an astigmatic CTF. The effective defocus at an arbitrary point g (scattering vector) in reciprocal space is defined by Equation 5. Adapted from Figure 3 of Mindell and Grigorieff (2003).
Figure 2:
Diagnostic image from micrograph #1 of set #7 of the CTF challenge, output by CTFFIND4 using runtime parameters detailed in Table 3. The 2-dimensional CTF (CTF fit) is overlayed onto the preprocessed amplitude spectrum (A d) up to the radius corresponding to g max.
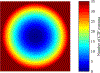
Figure 3:
Image E generated from the CTF fit to micrograph #1 of set #7 of the CTF challenge. At every pixel (corresponding to a spatial frequency vector g), this image records n, the number of preceding CTF extrema (Equation 11). Here this value is color-coded, so that pixels at spatial frequencies before the first extremum of the CTF, which have value 0, are displayed in dark blue. Pixels that have 35 or more preceding CTF extrema are shown in dark red.
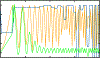
Figure 4:
Output diagnostic plots describing the experimental amplitudes (Ad1D, green), the fit CTF (CTFfit1D, orange) and goodness of fit (CC fit, blue) for micrograph #1 of set #7 of the CTF challenge. For this micrograph, the final estimates were Δf1=29070 Å, Δf2=28313Å and α_ast_ = 56.5°. The highest resolution at which Thon rings were deemed to be modeled correctly was 6.5 Å. The experimental amplitude profile (green) is normalized such that: the minima of the oscillations are set to 0.0; the second peak of the power spectrum (in this case at around 0.04) is 0.95; the maxima of oscillations are further normalized to 0.1 if their maxima would be <0.1 otherwise. Because of aliasing, one does not observe zeroes in CTFfit1D. One would normally solve this by increasing N d, but we restricted ourselves to previously-used parameter values for this experiment (see caption to Table 3 for more details).
Similar articles
- Visualization and quality assessment of the contrast transfer function estimation.
Sheth LK, Piotrowski AL, Voss NR. Sheth LK, et al. J Struct Biol. 2015 Nov;192(2):222-34. doi: 10.1016/j.jsb.2015.06.012. Epub 2015 Jun 12. J Struct Biol. 2015. PMID: 26080023 - FASTDEF: fast defocus and astigmatism estimation for high-throughput transmission electron microscopy.
Vargas J, Otón J, Marabini R, Jonic S, de la Rosa-Trevín JM, Carazo JM, Sorzano CO. Vargas J, et al. J Struct Biol. 2013 Feb;181(2):136-48. doi: 10.1016/j.jsb.2012.12.006. Epub 2012 Dec 20. J Struct Biol. 2013. PMID: 23261401 - Fast, robust, and accurate determination of transmission electron microscopy contrast transfer function.
Sorzano CO, Jonic S, Núñez-Ramírez R, Boisset N, Carazo JM. Sorzano CO, et al. J Struct Biol. 2007 Nov;160(2):249-62. doi: 10.1016/j.jsb.2007.08.013. Epub 2007 Aug 29. J Struct Biol. 2007. PMID: 17911028 - Role of optics in the accuracy of depth-from-defocus systems.
Blayvas I, Kimmel R, Rivlin E. Blayvas I, et al. J Opt Soc Am A Opt Image Sci Vis. 2007 Apr;24(4):967-72. doi: 10.1364/josaa.24.000967. J Opt Soc Am A Opt Image Sci Vis. 2007. PMID: 17361282 Review. - Theoretical aspects of image formation in the aberration-corrected electron microscope.
Rose H. Rose H. Ultramicroscopy. 2010 Apr;110(5):488-99. doi: 10.1016/j.ultramic.2009.10.003. Epub 2009 Oct 21. Ultramicroscopy. 2010. PMID: 19896274 Review.
Cited by
- Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Kocaman S, Lo YH, Krahn JM, Sobhany M, Dandey VP, Petrovich ML, Etigunta SK, Williams JG, Deterding LJ, Borgnia MJ, Stanley RE. Kocaman S, et al. PNAS Nexus. 2022 Jul 21;1(4):pgac118. doi: 10.1093/pnasnexus/pgac118. eCollection 2022 Sep. PNAS Nexus. 2022. PMID: 36090660 Free PMC article. - Heterogeneity in human hippocampal CaMKII transcripts reveals allosteric hub-dependent regulation.
Sloutsky R, Dziedzic N, Dunn MJ, Bates RM, Torres-Ocampo AP, Boopathy S, Page B, Weeks JG, Chao LH, Stratton MM. Sloutsky R, et al. Sci Signal. 2020 Jul 21;13(641):eaaz0240. doi: 10.1126/scisignal.aaz0240. Sci Signal. 2020. PMID: 32694170 Free PMC article. - Distinct conformational states of SARS-CoV-2 spike protein.
Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM Jr, Rawson S, Rits-Volloch S, Chen B. Cai Y, et al. Science. 2020 Sep 25;369(6511):1586-1592. doi: 10.1126/science.abd4251. Epub 2020 Jul 21. Science. 2020. PMID: 32694201 Free PMC article. - Structure of cyanobacterial photosystem I complexed with ferredoxin at 1.97 Å resolution.
Li J, Hamaoka N, Makino F, Kawamoto A, Lin Y, Rögner M, Nowaczyk MM, Lee YH, Namba K, Gerle C, Kurisu G. Li J, et al. Commun Biol. 2022 Sep 12;5(1):951. doi: 10.1038/s42003-022-03926-4. Commun Biol. 2022. PMID: 36097054 Free PMC article. - CryoEM structure and assembly mechanism of a bacterial virus genome gatekeeper.
Orlov I, Roche S, Brasilès S, Lukoyanova N, Vaney MC, Tavares P, Orlova EV. Orlov I, et al. Nat Commun. 2022 Nov 26;13(1):7283. doi: 10.1038/s41467-022-34999-8. Nat Commun. 2022. PMID: 36435855 Free PMC article.
References
- Fernando KV, Fuller SD, 2007. Determination of astigmatism in TEM images. J StructBiol 157, 189–200. - PubMed
- van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M,Patwardhan a., 2000. Singleparticle electron cryo-microscopy: towards atomic resolution. Quarterly reviews of biophysics 33, 307–69. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources