Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles - PubMed (original) (raw)
. 2015 Aug 17;210(4):529-39.
doi: 10.1083/jcb.201504117.
Geraldine Kong 1, Taro Mannen 2, Agata Sadowska 1, Simon Kobelke 1, Amanda Blythe 3, Gavin J Knott 3, K Swaminathan Iyer 3, Diwei Ho 3, Estella A Newcombe 4, Kana Hosoki 2, Naoki Goshima 5, Tetsuya Kawaguchi 2, Danny Hatters 4, Laura Trinkle-Mulcahy 6, Tetsuro Hirose 2, Charles S Bond 3, Archa H Fox 7
Affiliations
- PMID: 26283796
- PMCID: PMC4539981
- DOI: 10.1083/jcb.201504117
Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles
Sven Hennig et al. J Cell Biol. 2015.
Abstract
Prion-like domains (PLDs) are low complexity sequences found in RNA binding proteins associated with the neurodegenerative disorder amyotrophic lateral sclerosis. Recently, PLDs have been implicated in mediating gene regulation via liquid-phase transitions that drive ribonucleoprotein granule assembly. In this paper, we report many PLDs in proteins associated with paraspeckles, subnuclear bodies that form around long noncoding RNA. We mapped the interactome network of paraspeckle proteins, finding enrichment of PLDs. We show that one protein, RBM14, connects key paraspeckle subcomplexes via interactions mediated by its PLD. We further show that the RBM14 PLD, as well as the PLD of another essential paraspeckle protein, FUS, is required to rescue paraspeckle formation in cells in which their endogenous counterpart has been knocked down. Similar to FUS, the RBM14 PLD also forms hydrogels with amyloid-like properties. These results suggest a role for PLD-mediated liquid-phase transitions in paraspeckle formation, highlighting this nuclear body as an excellent model system for understanding the perturbation of such processes in neurodegeneration.
© 2015 Hennig et al.
Figures
Figure 1.
An interactome of paraspeckle proteins. (a) Schematic of the yeast two-hybrid mating strategy. (b) Example of a growth plate (left) with 48 mated yeast spots, each containing bait protein, with different candidate fusion proteins. The code of the grid position for each candidate is in
Table S1
. At right is the filter lift for the plate, color is β-galactosidase (β-Gal) activity. (c) Interactions between paraspeckle proteins, with numerical values binned into grayscale, as indicated. The values reflect both the strength of the interaction as well as the number of times it occurred in the two replicate experiments, see Materials and methods. (d) Example of a cotransformation with candidate proteins and negative controls. (e) Network diagram of the interactome, excluding putative paraspeckle proteins, with a cutoff of 8 for interaction (see Materials and methods), line thickness increasing with interaction score. Color coding is relevance to paraspeckle formation, determined by siRNA knockdown (Naganuma et al., 2012). Asterisks indicate proteins with PLDs (
Fig. S2
).
Figure 2.
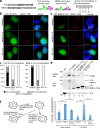
The RBM14 PLD mediates interaction with NONO and paraspeckle targeting. (a) Various truncations of RBM14 were tested for their ability to interact with NONO in the yeast two-hybrid assay, and the RBM14 PLD (residues 350–669) was the only fragment to recapitulate the interaction seen with full-length RBM14. (b) Western blotting for NONO showing that it is coimmunoprecipitated on GFP-trap resin from lysates containing YFP-RBM14 (lanes 2 and 5), or YFP-RBM14-PLD (lanes 3 and 6), but not YFP (lanes 1 and 4). Lysates in lanes 4–6 were treated with RNase A, which was effective at degrading RNA ∼30-fold over untreated samples, see Materials and methods. IP, immunoprecipitation; WB, Western blot. (c) Superresolution fluorescence micrograph of a HeLa nucleus: green, RBM14; red, NONO; blue, DAPI. Boxes are higher magnifications. Bars: (main image) 3 µm; (insets) 1 µm. (d) Fluorescence micrographs of representative HeLa cells transiently expressing YFP-RBM14 (top), YFP-RBM14-1-176 (middle), or YFP-RBM14-PLD (bottom). NEAT1 detected with FISH (red) to label paraspeckles. Green, YFP fluorescence; blue, DAPI. Both YFP-RBM14 and YFP-RBM14-PLD colocalize with paraspeckles, as indicated by yellow foci on overlays and arrows. Graph shows quantification of localization (see Materials and methods). Bars: (main images) 10 µm; (insets) 2 µm.
Figure 3.
The RBM14 PLD is essential for paraspeckle formation. (a) Schematics of secondary structure, consensus repeat motifs and mutational strategy for FUS and RBM14. ZNF, Zinc finger. (b and c) Fluorescence micrographs of representative HeLa cells transiently expressing YFP-NLS-FUS-PLD or YFP-RBM14-PLD (top), YFP-NLS-FUS-PLD partial Y→S mutant or YFP-RBM14-PLD partial Y→S mutant (middle), and YFP-NLS-FUS-PLD all Y→S mutant or YFP-RBM14-PLD All Y→S mutant (bottom). NEAT1 RNA detected with FISH (red) to label paraspeckles. Green, YFP fluorescence; blue, DAPI. The paraspeckle targeting by FUS or RBM14 PLD is lost when tyrosines are mutated. Bars, 10 µm (d) Quantification of colocalization for experiments in b and c, see Materials and methods. (e) Western blot for NONO showing it is coimmunoprecipitated on GFP-trap resin from lysates expressing YFP-RBM14 (lane 2) or YFP-RBM14-PLD (lane 3) but not YFP protein (lane 1) or either of the RBM14 PLD mutants (lanes 4 and 5). Bottom panels show anti-GFP Western of the same blot. WB, Western blot. (f) Schematic of paraspeckle rescue experiment and graph showing that transient, overexpressed, wild-type FUS or RBM14 can rescue paraspeckles after knockdown of endogenous FUS or RBM14, whereas the vector control or the Y→S mutant cannot. **, P < 0.02; means ± SD.
Figure 4.
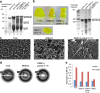
The RBM14 PLD forms a hydrogel with amyloid-like properties. (a) Coomassie blue staining of SDS-PAGE of purified recombinant proteins with evidence of some degradation for RBM14. GFP-FUS-PLD (lane 2), GFP-RBM14-PLD (lane 3), GFP-RBM14-PLD partial Y→S (lane 4), and GFP-RBM14-PLD All Y→S (lane 5) are shown. Size markers are shown in lane 1. (b) Photos of hydrogels formed by cooled, concentrated preparations of soluble GFP-FUS-PLD (left), GFP-RBM14-PLD (middle), and GFP-RBM14-PLD partial Y→S (right). The GFP-RBM14-PLD All Y→S was incapable of forming hydrogels (bottom). Bar, 2 mm. (right) Coomassie blue staining of hydrogel material, denatured and subject to SDS-PAGE, showing that hydrogels are enriched in full-length proteins. (c) Representative SEM images showing the fibrillar nature of hydrogels made with GFP-FUS-PLD (left), GFP-RBM14-PLD (middle), and GFP-RBM14-PLD partial Y→S (right). Bars, 200 nm. (d) X-ray diffraction of hydrogels made with GFP-FUS-PLD (left), GFP-RBM14-PLD (middle), and GFP-RBM14-PLD partial Y→S (right), showing the typical amyloid rings at 4.6 and 10 Å. (e) SDS solubility assay showing GFP-FUS-PLD, GFP-RBM14-PLD, or GFP-RBM14-PLD partial Y→S hydrogels are soluble in 2% SDS, whereas the pathological form (Htt46Q) of Huntingtin protein is not.
Comment in
- Paraspeckles: paragons of functional aggregation.
Courchaine E, Neugebauer KM. Courchaine E, et al. J Cell Biol. 2015 Aug 17;210(4):527-8. doi: 10.1083/jcb.201507052. J Cell Biol. 2015. PMID: 26283795 Free PMC article.
Similar articles
- Paraspeckles: paragons of functional aggregation.
Courchaine E, Neugebauer KM. Courchaine E, et al. J Cell Biol. 2015 Aug 17;210(4):527-8. doi: 10.1083/jcb.201507052. J Cell Biol. 2015. PMID: 26283795 Free PMC article. - Organization and function of paraspeckles.
Wang Y, Chen LL. Wang Y, et al. Essays Biochem. 2020 Dec 7;64(6):875-882. doi: 10.1042/EBC20200010. Essays Biochem. 2020. PMID: 32830222 Review. - The building process of the functional paraspeckle with long non-coding RNAs.
Yamazaki T, Hirose T. Yamazaki T, et al. Front Biosci (Elite Ed). 2015 Jan 1;7(1):1-41. doi: 10.2741/715. Front Biosci (Elite Ed). 2015. PMID: 25553361 Review. - Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles.
Hirose T, Yamazaki T, Nakagawa S. Hirose T, et al. Wiley Interdiscip Rev RNA. 2019 Nov;10(6):e1545. doi: 10.1002/wrna.1545. Epub 2019 May 1. Wiley Interdiscip Rev RNA. 2019. PMID: 31044562 Review. - Nuclear RNA foci from C9ORF72 expansion mutation form paraspeckle-like bodies.
Bajc Česnik A, Darovic S, Prpar Mihevc S, Štalekar M, Malnar M, Motaln H, Lee YB, Mazej J, Pohleven J, Grosch M, Modic M, Fonovič M, Turk B, Drukker M, Shaw CE, Rogelj B. Bajc Česnik A, et al. J Cell Sci. 2019 Mar 7;132(5):jcs224303. doi: 10.1242/jcs.224303. J Cell Sci. 2019. PMID: 30745340
Cited by
- Advances in the phase separation-organized membraneless organelles in cells: a narrative review.
Li W, Jiang C, Zhang E. Li W, et al. Transl Cancer Res. 2021 Nov;10(11):4929-4946. doi: 10.21037/tcr-21-1111. Transl Cancer Res. 2021. PMID: 35116344 Free PMC article. Review. - Prion-like domains drive CIZ1 assembly formation at the inactive X chromosome.
Sofi S, Williamson L, Turvey GL, Scoynes C, Hirst C, Godwin J, Brockdorff N, Ainscough J, Coverley D. Sofi S, et al. J Cell Biol. 2022 Apr 4;221(4):e202103185. doi: 10.1083/jcb.202103185. Epub 2022 Mar 15. J Cell Biol. 2022. PMID: 35289833 Free PMC article. - Blank spots on the map: some current questions on nuclear organization and genome architecture.
Adriaens C, Serebryannyy LA, Feric M, Schibler A, Meaburn KJ, Kubben N, Trzaskoma P, Shachar S, Vidak S, Finn EH, Sood V, Pegoraro G, Misteli T. Adriaens C, et al. Histochem Cell Biol. 2018 Dec;150(6):579-592. doi: 10.1007/s00418-018-1726-1. Epub 2018 Sep 20. Histochem Cell Biol. 2018. PMID: 30238154 Free PMC article. Review. - Aberrant Phase Transitions: Side Effects and Novel Therapeutic Strategies in Human Disease.
Verdile V, De Paola E, Paronetto MP. Verdile V, et al. Front Genet. 2019 Mar 22;10:173. doi: 10.3389/fgene.2019.00173. eCollection 2019. Front Genet. 2019. PMID: 30967892 Free PMC article. Review. - Matrin3: Disorder and ALS Pathogenesis.
Salem A, Wilson CJ, Rutledge BS, Dilliott A, Farhan S, Choy WY, Duennwald ML. Salem A, et al. Front Mol Biosci. 2022 Jan 10;8:794646. doi: 10.3389/fmolb.2021.794646. eCollection 2021. Front Mol Biosci. 2022. PMID: 35083279 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources