Evidence for a Common Origin of Blacksmiths and Cultivators in the Ethiopian Ari within the Last 4500 Years: Lessons for Clustering-Based Inference - PubMed (original) (raw)
Evidence for a Common Origin of Blacksmiths and Cultivators in the Ethiopian Ari within the Last 4500 Years: Lessons for Clustering-Based Inference
Lucy van Dorp et al. PLoS Genet. 2015.
Abstract
The Ari peoples of Ethiopia are comprised of different occupational groups that can be distinguished genetically, with Ari Cultivators and the socially marginalised Ari Blacksmiths recently shown to have a similar level of genetic differentiation between them (FST ≈ 0.023 - 0.04) as that observed among multiple ethnic groups sampled throughout Ethiopia. Anthropologists have proposed two competing theories to explain the origins of the Ari Blacksmiths as (i) remnants of a population that inhabited Ethiopia prior to the arrival of agriculturists (e.g. Cultivators), or (ii) relatively recently related to the Cultivators but presently marginalized in the community due to their trade. Two recent studies by different groups analysed genome-wide DNA from samples of Ari Blacksmiths and Cultivators and suggested that genetic patterns between the two groups were more consistent with model (i) and subsequent assimilation of the indigenous peoples into the expanding agriculturalist community. We analysed the same samples using approaches designed to attenuate signals of genetic differentiation that are attributable to allelic drift within a population. By doing so, we provide evidence that the genetic differences between Ari Blacksmiths and Cultivators can be entirely explained by bottleneck effects consistent with hypothesis (ii). This finding serves as both a cautionary tale about interpreting results from unsupervised clustering algorithms, and suggests that social constructions are contributing directly to genetic differentiation over a relatively short time period among previously genetically similar groups.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
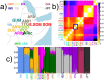
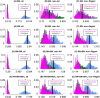
Fig 1. Description of sampled groups, ADMIXTURE clustering and F ST values.
(a) Geographic locations of sampled populations analysed, with the 12 Pagani populations [2] in larger font. The remaining 10 populations include the MKK and those from the 1000 Genomes Project (see
for details; CHI = CHB/CHS); locations on map for these 10 populations are indicative. All populations are colored by the group that many of their individuals were assigned to using fineSTRUCTURE; these 17 groups are referred to throughout using the label of one of the majority populations (see S3 Table). (b) Pairwise F ST comparing all groups (see S3 Table). (c) ADMIXTURE assuming 8 clusters applied to Pagani, 1KGP and MKK individuals, as labeled by the fineSTRUCTURE groups. In (b)-(c), the two Ari groups (ARIb, ARIc) are highlighted with the blue rectangle.
Fig 2. Full and simplified simulated histories under the marginalisation (MA) and remnants (RN) hypotheses.
History of populations simulated using MaCS [26]. (a) Thirteen populations simulated under the (i) Marginalisation (MA) model with Pop5 and Pop5b (representing the ARIc and ARIb, respectively) splitting at 20 gens, with a subsequent bottleneck in Pop5b, versus (ii) the Remnants model where Pop5b splits from Pop5/Pop6/Pop7 at 1700 gens and contributes migrants to Pop5/Pop6 between 200 and 300 gen ago. Otherwise all other groups and split times are the same between the two simulation scenarios. Orange arrows indicate migration from Pop10 into Pop5, Pop5b and Pop6. (b) Seven populations simulated under the Remnants model, with black arrows indicating migration from Pop5b into Pop5 and Pop4, and orange arrows indicating migration from Pop6 into Pop5, Pop5b and Pop6 with the given proportions. Pop5 and Pop5b split at varying times t ∈ {750, …, 1700}, with a bottleneck in Pop5b occurring 20–40 generations ago and the proportion of Pop5 comprised of Pop5b migrants varying from 50–90%.
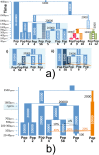
Fig 3. Inferred ancestry composition of groups under each analysis.
(top) Inferred ancestry composition of recipient groups when forming each group as mixtures of (a) all sampled groups, (b) all sampled groups except the Ari, and (c) all non-Pagani groups only. The colour of each group’s label provides the key for each pie, with Pagani groups geographically located on the map (roughly) according to the label most represented in the given group. All 1KGP groups and MKK are placed on the map loosely according to their relative geographic positions. (bottom) TVD XY values comparing the painting profiles for all pairwise comparisons of groups X, Y under each analysis, with scale at far right. Ari groups (ARIb/ARIc) are highlighted with black outlines in each plot.
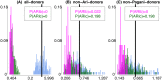
Fig 4. Differences in inferred ancestry under analyses (A)-(C) using F XY.
Differences in inferred ancestry under analyses (A)-(C) (using F XY; see Methods) between all pairings of ARIb individuals (pink), all pairings of ARIc individuals (green), and all pairings of one ARIb and one ARIc individual (cyan). In each plot the black vertical line gives the mean difference across the pairings of one ARIb and one ARIc, with P(ARIb), P(ARIc) giving the proportion of ARIb and ARIc pairings, respectively, with a difference greater than or equal to this mean.
Fig 5. Differences in inferred ancestry under analyses (A)-(C) using F XY applied to simulated data.
Differences in inferred ancestry under analyses (A)-(C) (using F XY; see Methods) between all pairings of simulated “ARIb” individuals (Pop5b, pink), all pairings of simulated “ARIc” individuals (Pop5, green), and all pairings of one “ARIb” and one “ARIc” individual (cyan), for the “MA”, “RN”, “RN+BN” and “RN+BN+80%” “full” simulations. In each plot the black vertical line gives the mean difference across the pairings of one Pop5b and one Pop5, with P(Pop5b), P(Pop5) giving the proportion of Pop5b and Pop5 pairings, respectively, with a difference greater than or equal to this mean.
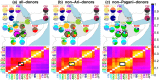
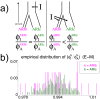
Fig 6. Effect on genetic similarity of two possible timings of DNA introgression.
(a) If the DNA introgression from source I occurred before the split of the Blacksmiths (ARIb) and Cultivators (ARIc) and subsequent genetic isolation of the Blacksmiths, as in the left depiction, the genetic similarity among ARIb for segments inherited from I (i.e. ϕIARIb) relative to the pre-introgression segments from the ancestral population A (ϕAARIb) should be the same as the analogous ratio of genetic similarity among the ARIc (i.e. ϕIARIc/ϕAARIc). In contrast, if the introgression from I occurred more recently than the split and isolation, as in the right depiction, the ratio (ϕIARIb/ϕAARIb) should be less than the ratio (ϕIARIc/ϕAARIc), since the component from I has experienced less drift effects than the component from A in the ARIb. (b) The distributions of (ϕIARIb/ϕAARIb) and (ϕIARIc/ϕAARIc) across all pairwise comparisons of individuals within each Ari group, when segments’ sources were inferred using the E-M model with a threshold of 0.94 when assuming I is the “West Eurasian” source (see Methods). The strong similarity in distributions is consistent with the introgression I occurring less recently than the split.
Similar articles
- Analysis of three RFLPs of the COL1A2 (Type I Collagen) in the Amhara and the Oromo of Ethiopia.
De Stefano GF, Martínez-Labarga C, Casalotti R, Tartaglia M, Novelletto A, Pepe G, Rickards O. De Stefano GF, et al. Ann Hum Biol. 2002 Jul-Aug;29(4):432-41. doi: 10.1080/03014460110101440. Ann Hum Biol. 2002. PMID: 12160476 - HLA class II allele and haplotype frequencies in Ethiopian Amhara and Oromo populations.
Fort M, de Stefano GF, Cambon-Thomsen A, Giraldo-Alvarez P, Dugoujon JM, Ohayon E, Scano G, Abbal M. Fort M, et al. Tissue Antigens. 1998 Apr;51(4 Pt 1):327-36. doi: 10.1111/j.1399-0039.1998.tb02971.x. Tissue Antigens. 1998. PMID: 9583804 - Clustering humans: on biological boundaries.
Lorusso L, Boniolo G. Lorusso L, et al. Stud Hist Philos Biol Biomed Sci. 2008 Mar;39(1):163-70. doi: 10.1016/j.shpsc.2007.12.005. Epub 2008 Feb 12. Stud Hist Philos Biol Biomed Sci. 2008. PMID: 18331963 - The use of racial, ethnic, and ancestral categories in human genetics research.
Race, Ethnicity, and Genetics Working Group. Race, Ethnicity, and Genetics Working Group. Am J Hum Genet. 2005 Oct;77(4):519-32. doi: 10.1086/491747. Epub 2005 Aug 29. Am J Hum Genet. 2005. PMID: 16175499 Free PMC article. Review.
Cited by
- Native American gene flow into Polynesia predating Easter Island settlement.
Ioannidis AG, Blanco-Portillo J, Sandoval K, Hagelberg E, Miquel-Poblete JF, Moreno-Mayar JV, Rodríguez-Rodríguez JE, Quinto-Cortés CD, Auckland K, Parks T, Robson K, Hill AVS, Avila-Arcos MC, Sockell A, Homburger JR, Wojcik GL, Barnes KC, Herrera L, Berríos S, Acuña M, Llop E, Eng C, Huntsman S, Burchard EG, Gignoux CR, Cifuentes L, Verdugo RA, Moraga M, Mentzer AJ, Bustamante CD, Moreno-Estrada A. Ioannidis AG, et al. Nature. 2020 Jul;583(7817):572-577. doi: 10.1038/s41586-020-2487-2. Epub 2020 Jul 8. Nature. 2020. PMID: 32641827 Free PMC article. - The Genetic Ancestry of Modern Indus Valley Populations from Northwest India.
Pathak AK, Kadian A, Kushniarevich A, Montinaro F, Mondal M, Ongaro L, Singh M, Kumar P, Rai N, Parik J, Metspalu E, Rootsi S, Pagani L, Kivisild T, Metspalu M, Chaubey G, Villems R. Pathak AK, et al. Am J Hum Genet. 2018 Dec 6;103(6):918-929. doi: 10.1016/j.ajhg.2018.10.022. Am J Hum Genet. 2018. PMID: 30526867 Free PMC article. - Changes in the fine-scale genetic structure of Finland through the 20th century.
Kerminen S, Cerioli N, Pacauskas D, Havulinna AS, Perola M, Jousilahti P, Salomaa V, Daly MJ, Vyas R, Ripatti S, Pirinen M. Kerminen S, et al. PLoS Genet. 2021 Mar 4;17(3):e1009347. doi: 10.1371/journal.pgen.1009347. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33661898 Free PMC article. - Hunter-gatherer genomes reveal diverse demographic trajectories during the rise of farming in Eastern Africa.
Gopalan S, Berl REW, Myrick JW, Garfield ZH, Reynolds AW, Bafens BK, Belbin G, Mastoras M, Williams C, Daya M, Negash AN, Feldman MW, Hewlett BS, Henn BM. Gopalan S, et al. Curr Biol. 2022 Apr 25;32(8):1852-1860.e5. doi: 10.1016/j.cub.2022.02.050. Epub 2022 Mar 9. Curr Biol. 2022. PMID: 35271793 Free PMC article. - A genetic chronology for the Indian Subcontinent points to heavily sex-biased dispersals.
Silva M, Oliveira M, Vieira D, Brandão A, Rito T, Pereira JB, Fraser RM, Hudson B, Gandini F, Edwards C, Pala M, Koch J, Wilson JF, Pereira L, Richards MB, Soares P. Silva M, et al. BMC Evol Biol. 2017 Mar 23;17(1):88. doi: 10.1186/s12862-017-0936-9. BMC Evol Biol. 2017. PMID: 28335724 Free PMC article.
References
- Pankhurst A (1999) ‘Caste’ in Africa: The Evidence from South-Western Ethiopia Reconsidered. Africa: 485–509.
- Freeman D, Pankhurst A (2003) Peripheral People: The Excluded Minorities of Ethiopia. London: Hurst and Company.
- Gebre Y (1995) The Ari of Southwestern Ethiopia: An Exploratory Study of Production Practices. Social Anthropology Dissertation Series.
- Biasutti R (1905) Pastori, agricoltori e cacciatori nell ’Africa Orientale’. Bolletino del Reale Societa geografica italiana 6: 155–179.
Publication types
MeSH terms
Grants and funding
- 098386/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- 098387/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- 098051/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 098386/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous