Architecture of the fungal nuclear pore inner ring complex - PubMed (original) (raw)
. 2015 Oct 2;350(6256):56-64.
doi: 10.1126/science.aac9176. Epub 2015 Aug 27.
Christopher J Bley 1, Karsten Thierbach 1, Stefan Petrovic 1, Sandra Schilbach 1, Daniel J Mayo 1, Thibaud Perriches 1, Emily J Rundlet 1, Young E Jeon 1, Leslie N Collins 1, Ferdinand M Huber 1, Daniel H Lin 1, Marcin Paduch 2, Akiko Koide 2, Vincent Lu 2, Jessica Fischer 3, Ed Hurt 3, Shohei Koide 2, Anthony A Kossiakoff 2, André Hoelz 4
Affiliations
- PMID: 26316600
- PMCID: PMC4826903
- DOI: 10.1126/science.aac9176
Architecture of the fungal nuclear pore inner ring complex
Tobias Stuwe et al. Science. 2015.
Abstract
The nuclear pore complex (NPC) constitutes the sole gateway for bidirectional nucleocytoplasmic transport. We present the reconstitution and interdisciplinary analyses of the ~425-kilodalton inner ring complex (IRC), which forms the central transport channel and diffusion barrier of the NPC, revealing its interaction network and equimolar stoichiometry. The Nsp1•Nup49•Nup57 channel nucleoporin heterotrimer (CNT) attaches to the IRC solely through the adaptor nucleoporin Nic96. The CNT•Nic96 structure reveals that Nic96 functions as an assembly sensor that recognizes the three-dimensional architecture of the CNT, thereby mediating the incorporation of a defined CNT state into the NPC. We propose that the IRC adopts a relatively rigid scaffold that recruits the CNT to primarily form the diffusion barrier of the NPC, rather than enabling channel dilation.
Copyright © 2015, American Association for the Advancement of Science.
Conflict of interest statement
The authors declare no financial conflicts of interest.
Figures
Fig. 1. Reconstitution and Dissection of the IRC
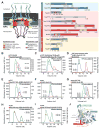
(A) Cross-sectional schematic representation of the NPC and domain structures of the Chaetomium thermophilum nucleoporins (nups). Black lines indicate regions used for reconstitution. U, unstructured; T, TAIL; NTD, N-terminal domain; MID, middle domain; SH3, Src-homology 3-like domain; L, linker domain; APD, auto-proteolytic domain; RRM, RNA recognition motif; M, membrane-binding motif; FG repeats, phenylalanine-glycine repeats; CCS, coiled-coil segment; α/β, α/β insertion domain; IRC, inner ring complex. (B to I) Pair-wise biochemical interaction analyses of various reconstituted nup complexes. Size-exclusion chromatography coupled to multiangle light scattering (SEC-MALS) profiles of nup or nup complexes are shown individually (red and blue) and after their preincubation (green). Measured molecular masses are indicated for the peak fractions. (J) Structure of the Nup82NTD•Nup159T•Nup145NAPD cytoplasmic filament nup complex (CFC) is shown in a cartoon representation. Nup145NAPD (green), Nup159T (red), and Nup82NTD (blue), the Nup82NTD helical 4CD (gray) and 6CD (orange) insertions and FGL loop (yellow), and the conserved Nup145NAPD K/R loop (purple) are illustrated.
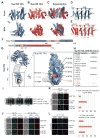
Fig. 2. Structural and functional analyses of the TAIL domains of Nup192 and Nup188
Cartoon representations of (A) Nup192TAIL (blue), (B) Nup188TAIL (red), and (C) a superposition of the two structures is shown in two different orientations. (D, E) Schematic representation of Nup192TAIL and Nup188TAIL structures. Positions of HEAT and ARM repeats are indicated. (F) Domain structures of Nup192 and Nic96 are shown; black bars indicate fragments used for interaction studies in (H). (G) Docking of Nup192TAIL and the previously determined Nup192NTD crystal structure into the yeast Nup192 EM envelope (8, 32). The Inset illustrates the position of Nup192TAIL and is expanded on the right, rotated by 90º. Surface representation of Nup192TAIL with the location of the analyzed mutations and their effect on Nic96R2 binding indicated; no effect (green), decreased binding (orange), abolished binding (red). (H) Summary of tested Nup192TAIL mutants and their effect on SUMO-Nic96R2 binding; (-) no effect, (+) decreased binding, (+++) abolished binding. (I) Growth analysis of S. cerevisiae strains carrying the indicated GFP-NUP192 variants. Serial dilutions of the respective cells were spotted onto YPD plates and grown for 2–3 days. (J) Subcellular localization of mCherry-Nup192 variants (red) and Nup57-GFP (green) in a _nup192Δ̃/NUP57-GFP_strain. (K) Subcellular localization of the 60S ribosomal export reporter Rpl25-mCherry (red) and GFP-tagged Nup192 variants (green) in a nup192Δ strain. Representative images and quantification of nuclear Rpl25-mCherry retention are shown on the right. (L) mRNA export assay in a nup192Δ strain carrying GFP-NUP192 variants. Representative images and quantification of nuclear poly(A)+ RNA retention are shown. Cells were analyzed at the indicated temperatures and incubation times. Error bars represent the standard deviation. Scalebars are 5 μm.
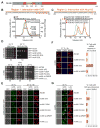
Fig. 3. Nic96 is an adaptor protein that attaches the CNT to Nup192
(A) Domain structure of Nic96 is shown. R1, region 1; R2, region 2. (B) The positions of the mutated leucine residues of the LLLL mutant are indicated below the primary Nic96R1 sequence. SEC-MALS profiles of CNT (black), Nic96R1 LLLL mutant (blue) and after preincubation of the CNT with wild type Nic96R1 (green) or Nic96R1 LLLL (red). Measured molecular masses are indicated for the peak fractions. (C) The positions of the mutated residues are indicated below the primary Nic96R2 sequence and colored according to their effect on Nup192TAIL binding; no effect (green), mild effect (orange), abolished binding (red). SEC profiles of Nup192TAIL preincubated with wild type Nic96R2 (green), Nic96R2 I294A (orange), and Nic96R2 F298A (red). (D) Growth analysis of S. cerevisiae strains carrying the indicated GFP-NIC96 variants. Serial dilutions of the respective cells were spotted onto indicated plates and grown for 3–5 days at the specified temperatures. (E) Subcellular localization of mCherry-Nic96 variants (red) and Nup57-GFP (green) in a nic96Δ̃/NUP57-GFP strain. (F) mRNA export assay in a nic96Δ̃NUP57 -GFP strain carrying GFP-NIC96 variants. Representative images and quantification of nuclear poly(A)+ RNA retention are shown. (G) Subcellular localization of the 60S ribosomal export reporter Rpl25-mCherry (red) and GFP-tagged Nup49 variants (green) in a nic96Δ strain. Quantification of nuclear Rpl25-mCherry retention is shown on the right. Cells were analyzed at the indicated temperatures and incubation times. Error bars represent the standard deviation. Scalebars are 5 μm.
Fig. 4. Crystal structure of the intact Nsp1•Nup49•Nup57 CNT bound to Nic96R1 and sAB-158
(A) Domain structures of the C. thermophilum channel nups Nsp1, Nup49, Nup57, the adaptor nup Nic96, and sAB-158. Black lines indicate the construct boundaries of the crystallized CNT•Nic96R1•sAB-158 complex. The constructs of the three channel nups comprise all regions with predicted secondary structure elements and only lack their unstructured N-terminal FG repeat regions. VH, heavy chain variable region; CH, heavy chain constant region; VL, light chain variable region; CL, light chain constant region. (B) Cartoon representation of the Nic96R1•CNT crystal structure viewed from three sides. For clarity, sAb-158 has been omitted. (C) Details of the Nic96R1-CNT interaction, illustrating the three channel nups and Nic96R1 in surface and cartoon representation, respectively. The Inset marks the region enlarged and 90º rotated in the middle panel. A further 90º rotated view is shown on the right. Nic96R1 (red) and Nup49 (magenta) residues that abolish CNT-Nic96R1 complex formation upon mutation (Figures 3 and 5) are indicated. (D) Structure of the CNT•Nic96R1•sAB-158 dimer shown in cartoon representation. The two copies of the CNT, Nic96R1, and sAB-158 are shown in blue/yellow, magenta/red, and gray, respectively. A 90º rotated view is shown on the right. The two CNT•Nic96R1•sAB-158 complexes in the asymmetric unit are related by two-fold rotational symmetry (black oval). Notably, the N-termini of all channel nups in the dimer point in the same direction, which would permit the FG repeats to project towards the central transport channel of the NPC.
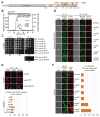
Fig. 5. The intact CNT is recruited to the NPC by its interaction with Nic96
(A) Domain structure of Nup49, indicating the corresponding EVPIP mutations (red) of the S. cerevisiae nup49-313 allele (37). (B) SEC-MALS profiles of the CNTEPP mutant (blue), Nic96R1-R2•Nup192TAIL (red) and after their preincubation (green). As reference, wild type CNT•Nic96R1-R2•Nup192TAIL is shown (black). Measured molecular masses are indicated for the peak fractions. (C) Growth analysis of S. cerevisiae strains carrying the indicated GFP-NUP49 variants. Serial dilutions of the respective cells were spotted onto YPD plates and grown for 2–4 days at the specified temperatures. (D) Subcellular localization of mCherry-Nup49 variants (red) and Nup57-GFP (green) in a _nup49Δ̃/NUP57-GFP_s strain. (E) mRNA export assay in a nup49Δ strain carrying GFP-NUP49 variants. Representative images and quantification of nuclear poly(A)+ RNA retention are shown. (F) Subcellular localization of the 60S ribosomal export reporter Rpl25-mCherry (red) and GFP-tagged Nup49 variants (green) in a nup49Δ strain. Quantification of nuclear Rpl25-mCherry retention is shown on the right. Cells were analyzed at the indicated temperatures and incubation times. Error bars represent the standard deviation. Scalebars are 5 μm.
Fig. 6. Proposed architecture of the NPC inner ring scaffold
Sixteen copies of the IRC are anchored to the nuclear pore membrane and arranged in a ring-shaped scaffold in an anti-parallel fashion to satisfy the established 8-fold and 2-fold symmetry axes of the NPC. Each IRC is composed of the channel nucleoporins Nsp1, Nup49, and Nup57 (CNT, red), the adaptor nucleoporins Nup192 (blue), Nic96 (yellow), Nup53 (magenta), and Nup145N (orange). The cytoplasmic and nuclear CNC rings (gray), the putative location of the POMs (green), and the inner ring (red) are shown. On the cytoplasmic side, Nup145N attaches the cytoplasmic filament nucleoporins Nup82 and Nup159 (CFC, cyan). The primarily unstructured adaptor nucleoporins Nup53 and Nup145N mediate the association of various structured IRC components and thus are critical for the IRC scaffold integrity. Nic96 functions as an assembly sensor that recognizes the conformation of the overall 4-shaped three-stranded coiled-coil domain architecture of the CNT, thereby mediating the selective incorporation of a defined CNT state into the NPC. The CNTs are positioned in the equatorial plane of the inner ring with the FG repeats of all three channel nups projecting towards the central transport channel. The anti-parallel orientation of adjacent IRCs would generate two CNT rings, one at the top and another at the bottom of the central transport channel. This arrangement would allow for the formation of a transport factor mediated FG repeat meshwork or hydrogel, which would further reinforce the inner ring scaffold. For size reference, a β-karyopherin•cargo complex is shown in gray.
Comment in
- STRUCTURAL BIOLOGY. Locking down the core of the pore.
Ullman KS, Powers MA. Ullman KS, et al. Science. 2015 Oct 2;350(6256):33-4. doi: 10.1126/science.aad3797. Science. 2015. PMID: 26430103 No abstract available.
Similar articles
- Structural basis of the nic96 subcomplex organization in the nuclear pore channel.
Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. Schrader N, et al. Mol Cell. 2008 Jan 18;29(1):46-55. doi: 10.1016/j.molcel.2007.10.022. Mol Cell. 2008. PMID: 18206968 - Architecture of the cytoplasmic face of the nuclear pore.
Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SG, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A. Bley CJ, et al. Science. 2022 Jun 10;376(6598):eabm9129. doi: 10.1126/science.abm9129. Epub 2022 Jun 10. Science. 2022. PMID: 35679405 Free PMC article. - STRUCTURAL BIOLOGY. Locking down the core of the pore.
Ullman KS, Powers MA. Ullman KS, et al. Science. 2015 Oct 2;350(6256):33-4. doi: 10.1126/science.aad3797. Science. 2015. PMID: 26430103 No abstract available. - Functional architecture of the nuclear pore complex.
Grossman E, Medalia O, Zwerger M. Grossman E, et al. Annu Rev Biophys. 2012;41:557-84. doi: 10.1146/annurev-biophys-050511-102328. Annu Rev Biophys. 2012. PMID: 22577827 Review. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review.
Cited by
- Affinity Isolation of Endogenous Saccharomyces Cerevisiae Nuclear Pore Complexes.
Nudelman I, Fernandez-Martinez J, Rout MP. Nudelman I, et al. Methods Mol Biol. 2022;2502:3-34. doi: 10.1007/978-1-0716-2337-4_1. Methods Mol Biol. 2022. PMID: 35412228 Free PMC article. - Structure of cytoplasmic ring of nuclear pore complex by integrative cryo-EM and AlphaFold.
Fontana P, Dong Y, Pi X, Tong AB, Hecksel CW, Wang L, Fu TM, Bustamante C, Wu H. Fontana P, et al. Science. 2022 Jun 10;376(6598):eabm9326. doi: 10.1126/science.abm9326. Epub 2022 Jun 10. Science. 2022. PMID: 35679401 Free PMC article. - Evolutionary divergence of the nuclear pore complex from fungi to metazoans.
Chopra K, Bawaria S, Chauhan R. Chopra K, et al. Protein Sci. 2019 Mar;28(3):571-586. doi: 10.1002/pro.3558. Epub 2018 Dec 24. Protein Sci. 2019. PMID: 30488506 Free PMC article. - The Great Escape: mRNA Export through the Nuclear Pore Complex.
De Magistris P. De Magistris P. Int J Mol Sci. 2021 Oct 29;22(21):11767. doi: 10.3390/ijms222111767. Int J Mol Sci. 2021. PMID: 34769195 Free PMC article. Review. - Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex.
Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, Weis K. Onischenko E, et al. Cell. 2017 Nov 2;171(4):904-917.e19. doi: 10.1016/j.cell.2017.09.033. Epub 2017 Oct 12. Cell. 2017. PMID: 29033133 Free PMC article.
References
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. - PubMed
- Bui KH, von Appen A, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. - PubMed
- Eisenhardt N, Redolfi J, Antonin W. Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J Cell Sci. 2014;127:908–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01-GM094588/GM/NIGMS NIH HHS/United States
- T32 GM007616/GM/NIGMS NIH HHS/United States
- R01 GM090324/GM/NIGMS NIH HHS/United States
- U01 GM094588/GM/NIGMS NIH HHS/United States
- P30 CA014599/CA/NCI NIH HHS/United States
- ACB-12002/PHS HHS/United States
- AGM-12006/PHS HHS/United States
- R01-GM111461/GM/NIGMS NIH HHS/United States
- U54 GM087519/GM/NIGMS NIH HHS/United States
- R01 GM117360/GM/NIGMS NIH HHS/United States
- P30-CA014599/CA/NCI NIH HHS/United States
- U54-GM087519/GM/NIGMS NIH HHS/United States
- R01-GM090324/GM/NIGMS NIH HHS/United States
- R01 GM111461/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources