Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and prioritization for discovery - PubMed (original) (raw)
Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and prioritization for discovery
Jeffrey D Rudolf et al. J Ind Microbiol Biotechnol. 2016 Mar.
Abstract
The enediynes are one of the most fascinating families of bacterial natural products given their unprecedented molecular architecture and extraordinary cytotoxicity. Enediynes are rare with only 11 structurally characterized members and four additional members isolated in their cycloaromatized form. Recent advances in DNA sequencing have resulted in an explosion of microbial genomes. A virtual survey of the GenBank and JGI genome databases revealed 87 enediyne biosynthetic gene clusters from 78 bacteria strains, implying that enediynes are more common than previously thought. Here we report the construction and analysis of an enediyne genome neighborhood network (GNN) as a high-throughput approach to analyze secondary metabolite gene clusters. Analysis of the enediyne GNN facilitated rapid gene cluster annotation, revealed genetic trends in enediyne biosynthetic gene clusters resulting in a simple prediction scheme to determine 9- versus 10-membered enediyne gene clusters, and supported a genomic-based strain prioritization method for enediyne discovery.
Keywords: Biosynthetic gene cluster; Enediyne polyketide synthase; Genome mining; Genome neighborhood network; Natural products.
Figures
Fig. 1
Structures of the 11 known enediynes, (A) five 9-membered and (B) six 10-membered representatives, with their enediyne cores highlighted in red. The sporolides, cyanosporasides, and fijolides were proposed to be derived from 9-membered enediynes. Given in parentheses are the years when each enediyne structure was established.
Fig. 2
Characterization of the enediyne biosynthetic machinery providing an opportunity to explore bacterial genomes for the discovery of new enediyne natural products. (A) Ten known enediyne clusters highlighting the pksE cassettes (i.e., E3, E4, E5, E, E10, red genes) common to all enediynes and used as a beacon for enediyne producers. The surrounding ORFs in the gene clusters, forming the pksE genome neighborhoods, are color-coded to signify their involvement in peripheral moiety biosynthesis, pathway regulation, and self-resistance. (B) Unified model for enediyne core biosynthesis. Conserved proteins in all enediynes (E, E3, E4, E5, and E10) produces an ACP-tethered or free polyketide intermediate that is converted into the proposed enediyne cores in the presence of 9- (path a) or 10-membered (path b) associated enzymes. Examples of 9- and 10-membered associated enzymes are named according to the homologues of C-1027 and CAL, respectively.
Fig. 3
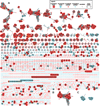
Genome neighborhood network (GNN) for the enediyne family of natural products. (A) The GNN displayed with an E-value threshold of 10−8. Each node is colored and shaped based on the enediyne it produces, labeled with its corresponding gene name or ORF number, and highlighted if it has been functionally characterized (see inset legend). (B–D) Selected families of conserved proteins involved in both 9- and 10-membered, only 9-membered, or only 10-membered enediyne biosynthesis, respectively. (E) Members of the sequestration apoprotein family for 9-membered enediynes and the self-sacrifice protein family for CAL.
Fig. 4
GNN depicting genera of enediyne producers. The GNN displayed with an E-value threshold of 10−8. Each node is colored based on the taxonomic identification of the strain it is found in, shaped based on the size of the enediyne core it produces, labeled with its corresponding gene name or ORF number, and highlighted if it has been functionally characterized (see inset legend).
Fig. 5
GNN depicting genetic location in reference to pksE. The GNN displayed with an E-value threshold of 10−8. Each node is colored based on its genetic distance from its pksE, shaped based on the size of the enediyne core it produces, labeled with its corresponding gene name or ORF number, and highlighted if it has been functionally characterized (see inset legend).
Fig. 6
High stringency GNN for the enediyne family of natural products. (A) The GNN displayed with an E-value threshold of 10−75. Each node is colored and shaped based on the enediyne it produces, labeled with its corresponding gene name or ORF number, and highlighted if it has been functionally characterized (see inset legend). For comparisons of GNNs at a lower threshold or with 9- or 10-membered enediyne prediction, see Figs. 3 or 7, respectively. (B) The E2 and E3 family is separated into three groups (see Fig. 3B for comparison at an E-value threshold of 10−8). (C) Families of conserved proteins (i.e., CalR3 and CalT6/T7) from gene clusters not containing an E2 homologue.
Fig. 7
Genome neighborhood network (GNN) facilitates the prediction of 9- and 10-membered enediynes. (A) The GNN displayed with an E-value threshold of 10−75. Using the E2 and CalR3 families for 9- and 10-membered enediyne indicators, respectively, each node is colored based and shaped on the size of the enediyne core it produces or is predicted to produce (see inset legend). Each node is labeled with its corresponding gene name or ORF number, and highlighted if it has been functionally characterized. See Fig. 6 for a complementary GNN colored based on the known enediyne it produces. See Fig. S2 for a GNN displaying the predicted 9- and 10-membered enediynes at an E-value threshold of 10−8. (B) The E2 and (C) CalR3 families as 9- and 10-membered enediyne indicators, respectively.
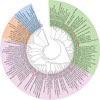
Fig. 8
Phylogenetic analysis of the E2 and E3 family of proteins conserved in enediyne gene clusters. The E2 and E3 proteins are separated into four groups: E2s (green), typical E3s with close relationships to the E2 family (pink), atypical E3s from nonactinobacteria (orange), and atypical E3s from actinobacteria (blue). DynU15 is phylogenetically distinct. Each protein is labeled with its locus tag with its corresponding bacteria strain in parentheses. The E2 and E3 proteins from known 9-membered enediyne gene clusters are represented with red dots and red diamonds, respectively. The E3 proteins from known 10-membered enediyne gene clusters are represented with purple diamonds. E3 proteins with E2 or CalR3 homologues in their gene clusters are highlighted with red stars or blue triangles, respectively. SgcE5 was used as an outgroup.
Similar articles
- Genome mining for new enediyne antibiotics.
Han EJ, Seyedsayamdost MR. Han EJ, et al. Curr Opin Chem Biol. 2024 Aug;81:102481. doi: 10.1016/j.cbpa.2024.102481. Epub 2024 Jun 24. Curr Opin Chem Biol. 2024. PMID: 38917732 Review. - Enediyne natural product biosynthesis unified by a diiodotetrayne intermediate.
Gui C, Kalkreuter E, Lauterbach L, Yang D, Shen B. Gui C, et al. Nat Chem Biol. 2024 Sep;20(9):1210-1219. doi: 10.1038/s41589-024-01636-y. Epub 2024 Jun 3. Nat Chem Biol. 2024. PMID: 38831037 - Strain Prioritization and Genome Mining for Enediyne Natural Products.
Yan X, Ge H, Huang T, Hindra, Yang D, Teng Q, Crnovčić I, Li X, Rudolf JD, Lohman JR, Gansemans Y, Zhu X, Huang Y, Zhao LX, Jiang Y, Van Nieuwerburgh F, Rader C, Duan Y, Shen B. Yan X, et al. mBio. 2016 Dec 20;7(6):e02104-16. doi: 10.1128/mBio.02104-16. mBio. 2016. PMID: 27999165 Free PMC article. - Iterative type I polyketide synthases involved in enediyne natural product biosynthesis.
Chen X, Ji R, Jiang X, Yang R, Liu F, Xin Y. Chen X, et al. IUBMB Life. 2014 Sep;66(9):587-95. doi: 10.1002/iub.1316. Epub 2014 Oct 3. IUBMB Life. 2014. PMID: 25278375 Review. - Bioactivity-guided genome mining reveals the lomaiviticin biosynthetic gene cluster in Salinispora tropica.
Kersten RD, Lane AL, Nett M, Richter TK, Duggan BM, Dorrestein PC, Moore BS. Kersten RD, et al. Chembiochem. 2013 May 27;14(8):955-62. doi: 10.1002/cbic.201300147. Epub 2013 May 3. Chembiochem. 2013. PMID: 23649992 Free PMC article.
Cited by
- Primed for Discovery.
Walker AS, Clardy J. Walker AS, et al. Biochemistry. 2024 Nov 5;63(21):2705-2713. doi: 10.1021/acs.biochem.4c00464. Epub 2024 Oct 15. Biochemistry. 2024. PMID: 39497571 Free PMC article. Review. - Genome mining for new enediyne antibiotics.
Han EJ, Seyedsayamdost MR. Han EJ, et al. Curr Opin Chem Biol. 2024 Aug;81:102481. doi: 10.1016/j.cbpa.2024.102481. Epub 2024 Jun 24. Curr Opin Chem Biol. 2024. PMID: 38917732 Review. - Enediyne natural product biosynthesis unified by a diiodotetrayne intermediate.
Gui C, Kalkreuter E, Lauterbach L, Yang D, Shen B. Gui C, et al. Nat Chem Biol. 2024 Sep;20(9):1210-1219. doi: 10.1038/s41589-024-01636-y. Epub 2024 Jun 3. Nat Chem Biol. 2024. PMID: 38831037 - Pseudogenomic insights into the evolution of Mycobacterium ulcerans.
Kyei-Baffour ES, Owusu-Boateng K, Isawumi A, Mosi L. Kyei-Baffour ES, et al. BMC Genomics. 2024 Jan 22;25(1):87. doi: 10.1186/s12864-024-10001-1. BMC Genomics. 2024. PMID: 38253991 Free PMC article. - Targeted Discovery of Cryptic Enediyne Natural Products via FRET-Coupled High-Throughput Elicitor Screening.
Han EJ, Lee SR, Townsend CA, Seyedsayamdost MR. Han EJ, et al. ACS Chem Biol. 2023 Aug 18;18(8):1854-1862. doi: 10.1021/acschembio.3c00281. Epub 2023 Jul 18. ACS Chem Biol. 2023. PMID: 37463302 Free PMC article.
References
- Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. - PubMed
- Boghaert ER, Sridharan L, Armellino DC, Khandke KM, DiJoseph JF, Kunz A, Dougher MM, Jiang F, Kalyandrug LB, Hamann PR, Frost P, Damle NK. Antibody-targeted chemotherapy with the calicheamicin conjugate hu3S193-N-acetyl γ calicheamicin dimethyl hydrazide targets Lewisy and eliminates Lewisy-positive human carcinoma cells and xenografts. Clin Cancer Res. 2004;10:4538–4549. - PubMed
- Buchanan GO, Williams PG, Feling RH, Kauffman CA, Jensen PR, Fenical W. Sporolides A and B: structurally unprecedented halogenated macrolides from the marine actinomycete Salinispora tropica. Org Lett. 2005;7:2731–2734. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources