Recent events dominate interdomain lateral gene transfers between prokaryotes and eukaryotes and, with the exception of endosymbiotic gene transfers, few ancient transfer events persist - PubMed (original) (raw)
Recent events dominate interdomain lateral gene transfers between prokaryotes and eukaryotes and, with the exception of endosymbiotic gene transfers, few ancient transfer events persist
Laura A Katz. Philos Trans R Soc Lond B Biol Sci. 2015.
Abstract
While there is compelling evidence for the impact of endosymbiotic gene transfer (EGT; transfer from either mitochondrion or chloroplast to the nucleus) on genome evolution in eukaryotes, the role of interdomain transfer from bacteria and/or archaea (i.e. prokaryotes) is less clear. Lateral gene transfers (LGTs) have been argued to be potential sources of phylogenetic information, particularly for reconstructing deep nodes that are difficult to recover with traditional phylogenetic methods. We sought to identify interdomain LGTs by using a phylogenomic pipeline that generated 13 465 single gene trees and included up to 487 eukaryotes, 303 bacteria and 118 archaea. Our goals include searching for LGTs that unite major eukaryotic clades, and describing the relative contributions of LGT and EGT across the eukaryotic tree of life. Given the difficulties in interpreting single gene trees that aim to capture the approximately 1.8 billion years of eukaryotic evolution, we focus on presence-absence data to identify interdomain transfer events. Specifically, we identify 1138 genes found only in prokaryotes and representatives of three or fewer major clades of eukaryotes (e.g. Amoebozoa, Archaeplastida, Excavata, Opisthokonta, SAR and orphan lineages). The majority of these genes have phylogenetic patterns that are consistent with recent interdomain LGTs and, with the notable exception of EGTs involving photosynthetic eukaryotes, we detect few ancient interdomain LGTs. These analyses suggest that LGTs have probably occurred throughout the history of eukaryotes, but that ancient events are not maintained unless they are associated with endosymbiotic gene transfer among photosynthetic lineages.
Keywords: endosymbiotic gene transfer; eukaryotic tree of life; horizontal gene transfer; phylogenomics.
© 2015 The Author(s).
Figures
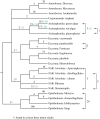
Figure 1.
An example of a recent interdomain LGT from prokaryotes to one minor clade (Metazoa) in one major clade (Opisthokonta) of eukaryotes. This tree exemplifies the many recent (e.g. 1MC1mc) interdomain transfers detected in this study (table 2 and figure 3). Abbreviations of taxa are as in table 1 and electronic supplementary material, S1, and the number following each name is a unique identifier from either OrthoMCL or GenBank. Analyses of this gene used PROTGAMMA, the best-fitting LG model and default parameters as implemented in R
ax
ML [49,50]. Most nodes are poorly supported and only bootstrap values above 80% are shown. Monophyletic clades are marked with solid lines, whereas the complex relationships among prokaryotes in the dashed clades probably represent a combination of poorly resolved phylogeny, LGT among prokaryotes and gene loss.
Figure 2.
Recent LGT events mapped onto representative lineages from the eukaryotic tree of life. Numbers at nodes represent the LGT events in table 3, and the synthetic tree is arbitrarily rooted on Opisthokonta. Numbers marked by asterisk are found in at least three minor clades within major clades and may represent synapomorphies for major clades. Arrows on the right mark shared putative LGTs found between non-sister minor clades. Numbers in green (grey) in parentheses are genes where eukaryotes fall sister to cyanobacteria and are hence putative EGTs. For simplicity, only a subset of lineages are included here and full taxonomic distributions can be found in the electronic supplementary material, tables S1 and S2. (Online version in colour.)
Figure 3.
Significant networks among lineages as determined by COPAP [54] based on presence–absence of LGTs. The green (dark) taxa are predominantly photosynthetic, and networks involving these minor clades indicate the potential influence of EGT on photosynthetic lineages. There is also a significant relationship between Entamoeba spp. (Am_ar) and parabasalids (Ex_pa) as has been previously observed [,–57]. The linking of fungi (Op_fu) and microbial opisthokonts (Op_ot; other Opisthokonta = Ichthyosporea plus lineages that are incertae sedis) probably represents shared retention of LGT events. (Online vesion in colour.)
Similar articles
- Old genes in new places: A taxon-rich analysis of interdomain lateral gene transfer events.
Cote-L'Heureux A, Maurer-Alcalá XX, Katz LA. Cote-L'Heureux A, et al. PLoS Genet. 2022 Jun 22;18(6):e1010239. doi: 10.1371/journal.pgen.1010239. eCollection 2022 Jun. PLoS Genet. 2022. PMID: 35731825 Free PMC article. - Assessing the effects of a sequestered germline on interdomain lateral gene transfer in Metazoa.
Jensen L, Grant JR, Laughinghouse HD 4th, Katz LA. Jensen L, et al. Evolution. 2016 Jun;70(6):1322-33. doi: 10.1111/evo.12935. Epub 2016 May 30. Evolution. 2016. PMID: 27139503 - Phylogenomic study indicates widespread lateral gene transfer in Entamoeba and suggests a past intimate relationship with parabasalids.
Grant JR, Katz LA. Grant JR, et al. Genome Biol Evol. 2014 Sep;6(9):2350-60. doi: 10.1093/gbe/evu179. Genome Biol Evol. 2014. PMID: 25146649 Free PMC article. - Lateral gene transfers and the evolution of eukaryotes: theories and data.
Katz LA. Katz LA. Int J Syst Evol Microbiol. 2002 Sep;52(Pt 5):1893-1900. doi: 10.1099/00207713-52-5-1893. Int J Syst Evol Microbiol. 2002. PMID: 12361302 Review. - Demystifying Eukaryote Lateral Gene Transfer (Response to Martin 2017 DOI: 10.1002/bies.201700115).
Leger MM, Eme L, Stairs CW, Roger AJ. Leger MM, et al. Bioessays. 2018 May;40(5):e1700242. doi: 10.1002/bies.201700242. Epub 2018 Mar 15. Bioessays. 2018. PMID: 29543982 Review.
Cited by
- Fungal evolution: cellular, genomic and metabolic complexity.
Naranjo-Ortiz MA, Gabaldón T. Naranjo-Ortiz MA, et al. Biol Rev Camb Philos Soc. 2020 Oct;95(5):1198-1232. doi: 10.1111/brv.12605. Epub 2020 Apr 17. Biol Rev Camb Philos Soc. 2020. PMID: 32301582 Free PMC article. - Microevolution, speciation and macroevolution in rhizobia: Genomic mechanisms and selective patterns.
Provorov NA, Andronov EE, Kimeklis AK, Onishchuk OP, Igolkina AA, Karasev ES. Provorov NA, et al. Front Plant Sci. 2022 Oct 25;13:1026943. doi: 10.3389/fpls.2022.1026943. eCollection 2022. Front Plant Sci. 2022. PMID: 36388581 Free PMC article. Review. - Gene refashioning through innovative shifting of reading frames in mosses.
Guan Y, Liu L, Wang Q, Zhao J, Li P, Hu J, Yang Z, Running MP, Sun H, Huang J. Guan Y, et al. Nat Commun. 2018 Apr 19;9(1):1555. doi: 10.1038/s41467-018-04025-x. Nat Commun. 2018. PMID: 29674719 Free PMC article. - Evolving Perspective on the Origin and Diversification of Cellular Life and the Virosphere.
Spang A, Mahendrarajah TA, Offre P, Stairs CW. Spang A, et al. Genome Biol Evol. 2022 May 31;14(6):evac034. doi: 10.1093/gbe/evac034. Genome Biol Evol. 2022. PMID: 35218347 Free PMC article. Review. - The plastid genome of some eustigmatophyte algae harbours a bacteria-derived six-gene cluster for biosynthesis of a novel secondary metabolite.
Yurchenko T, Ševčíková T, Strnad H, Butenko A, Eliáš M. Yurchenko T, et al. Open Biol. 2016 Nov;6(11):160249. doi: 10.1098/rsob.160249. Open Biol. 2016. PMID: 27906133 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous