The Proteome of Native Adult Müller Glial Cells From Murine Retina - PubMed (original) (raw)
The Proteome of Native Adult Müller Glial Cells From Murine Retina
Antje Grosche et al. Mol Cell Proteomics. 2016 Feb.
Abstract
To date, the proteomic profiling of Müller cells, the dominant macroglia of the retina, has been hampered because of the absence of suitable enrichment methods. We established a novel protocol to isolate native, intact Müller cells from adult murine retinae at excellent purity which retain in situ morphology and are well suited for proteomic analyses. Two different strategies of sample preparation - an in StageTips (iST) and a subcellular fractionation approach including cell surface protein profiling were used for quantitative liquid chromatography-mass spectrometry (LC-MSMS) comparing Müller cell-enriched to depleted neuronal fractions. Pathway enrichment analyses on both data sets enabled us to identify Müller cell-specific functions which included focal adhesion kinase signaling, signal transduction mediated by calcium as second messenger, transmembrane neurotransmitter transport and antioxidant activity. Pathways associated with RNA processing, cellular respiration and phototransduction were enriched in the neuronal subpopulation. Proteomic results were validated for selected Müller cell genes by quantitative real time PCR, confirming the high expression levels of numerous members of the angiogenic and anti-inflammatory annexins and antioxidant enzymes (e.g. paraoxonase 2, peroxiredoxin 1, 4 and 6). Finally, the significant enrichment of antioxidant proteins in Müller cells was confirmed by measurements on vital retinal cells using the oxidative stress indicator CM-H2DCFDA. In contrast to photoreceptors or bipolar cells, Müller cells were most efficiently protected against H2O2-induced reactive oxygen species formation, which is in line with the protein repertoire identified in the proteomic profiling. Our novel approach to isolate intact glial cells from adult retina in combination with proteomic profiling enabled the identification of novel Müller glia specific proteins, which were validated as markers and for their functional impact in glial physiology. This provides the basis to allow the discovery of novel glial specializations and will enable us to elucidate the role of Müller cells in retinal pathologies - a topic still controversially discussed.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
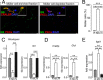
Fig. 1.
Itgb1-MACS allows efficient enrichment of morphologically intact Müller glia cells from murine retinae. A, Glutamine synthetase (GS) and cellular retinaldehyde binding protein (CRALBP) served as markers for Müller cells, PKCα labeling delineates rod photoreceptor bipolar cells (arrowhead in the inset marks a bipolar cell at higher magnification) and DAPI stains all nuclei in the sample. Note the excellent preservation of Müller cell morphology also depicted in the insert showing an enlarged view of an isolated Müller cell. Photoreceptor nuclei can easily discerned from that of all other cell types owing to their unique chromatin structure with tightly packed central chromatin core (arrows). Scale bars, 25 μm. B, The percentage of Müller cells in each cellular subpopulation was determined on the basis of the immunolabelings. The number of cells positive for the Müller cell marker glutamine synthetase was put into relation to the total cell number (number of DAPI-positive cell nuclei). Each bar represents values from 8 independent experiments for each of which retinae from two animals were pooled. ***p < 0.001. C–E, Quantitative RT-PCR experiments on cDNA transcribed from total RNA isolated from whole retinae (R), Müller cell enriched (MC) and Müller cell-depleted neuronal (N) cell subpopulations. C, Photoreceptor-specific transcripts rhodopsin and Nrl were present to a significantly lesser extent in the Müller cell-enriched samples than in whole retinal or Müller cell-depleted neuronal samples. D, Müller cell-specific transcripts (Cralbp and glutamine synthetase (Glul)) were strongly enriched in the Müller cell fraction and almost completely absent in the Müller cell-depleted fraction. E, Integrin β1 (Itgb1) is expressed at significantly higher levels in Müller glia than in retinal neurons. B–D, Values are given as ±S.E. and include results from five independent qRT-PCR experiments (n = 5). For sample generation cells from four retinae derived from two animals were pooled. ***p < 0.001, **p < 0.01, *p < 0.05.
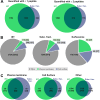
Fig. 2.
Total protein identifications from samples prepared by iST and subcellular fractionations. A, Numbers of protein identifications from iST (light green) versus subcellular fractionation method (blue) including one peptide hits (left venn diagram) and excluding one peptide hits (right venn diagram). Numbers of overlapping IDs are also indicated (dark green). B, All identified proteins from both fractionation approaches (values in brackets indicate proteins quantified with two or more peptides) are analyzed for subcellular localization by prediction of signal peptides for secretion and number of transmembrane domains. Protein IDs are categorized into: “Plasma Membrane” if a secretion signal peptide and at least one transmembrane domain were predicted (green), “Cell surface” if only a secretion peptide, but no transmembrane domain was predicted (blue). All other proteins are included under “Other” (gray). C, Identified protein numbers approaches (values in brackets indicate proteins quantified with two or more peptides) allocated to the three localization categories “Plasma Membrane,”, “Cell surface,” and “Other” are compared between the iST fractionation (green) and the subcellular fractionation (blue).
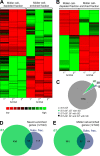
Fig. 3.
Comparison of protein identifications from Müller cell-enriched and -depleted samples. A, B, Heatmaps of hierarchical cluster analysis of significantly differently expressed proteins in Müller cell-enriched and -depleted fractions. Proteins and samples were clustered based on the respective protein abundances applying hierarchical clustering based on Euclidian distances. The corresponding heatmap is shown, with highly abundant proteins presented in green and low abundant proteins in red in the respective samples of (A) the iST approach or (B) the subcellular fractionation. C, Here we concentrated on proteins identified by both approaches (iST, subcellular fractionation (SF)) and determined whether the enrichment profiles of each protein found in the two approaches were in agreement with each other. For the vast majority of proteins, both approaches yielded a similar enrichment pattern (gray). Additionally, there were few proteins for which only one of the methods determined a significant difference (S.D.) in the expression level comparing both cell populations and the other method implicated an inverse enrichment, but did not reach significance (green, purple). Only one protein (black) was found to be significantly in enriched in Müller cells with one approach and the other approach indicated a significant enrichment in the neuronal fraction. D, E, Venn diagrams illustrating the overlapping and unique proteins identified with iST and subcellular fractionation of the Müller cell-depleted (neuronal) fraction (D) and the Müller cell enriched fraction (E). Threshold for enrichment to a specific fraction were set to ≥twofold enrichment and p < 0.05 (ANOVA of normalized cumulative peptide abundances).
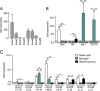
Fig. 4.
Quantitative RT-PCR (qRT-PCR) validation of selected anti-inflammatory genes identified as enriched in Müller cells. A, The general expression levels (mRNA) of the chosen anti-inflammatory genes in whole retinal extracts were determined to allow an estimation of the putative relevance of the respective gene in retinal physiology. Values represent the mean ± S.E. from three independent experiments. B, To obtain information about the annexin expression in microglia, we isolated this cell type in an additional enrichment step from the same retinal cell suspension from which subsequently also the Müller cells were isolated. Quantitative expression analysis using qRT-PCR confirms high purity of Müller and microglial fractions as the Müller cell specific glutamine synthetase (Glul) is highest expressed in the Müller cell fraction, whereas Nrl (photoreceptor marker) is primarily expressed in the neuronal fraction and the microglial markers Iba1 and CD11b are exclusively expressed in the CD11b-positive microglial fraction. Values are derived from four independent experiments for each analysis with retinae from two mice pooled per analysis. C, Expression analyses of annexin 1–8 at mRNA level in Müller cells compared with retinal neurons largely mirror the results obtained by quantitative proteomic analysis. Of note, the Müller cell-specific expression of all genes appears to be more pronounced at protein levels (for comparison the respective factor of enrichment taken from the proteomic analysis is given in the brackets below the gene symbol; annexin 8 (Anxa8) was not detected in our proteomic analyses, n.f., not found). Only Anxa3 showed highest expression levels in microglia, whereas most of the other annexins were primarily expressed in Müller cells (Anxa1, Anxa2, Anxa4, Anxa5). Values represent the mean ± S.E. from 5–6 independent experiments. For each experiment, cells were pooled from 4 retinae originating from 2 animals. B, C, **p < 0.01, *p < 0.05; significantly different expression level of the gene in the respective sample compared with whole retinal extracts: ○○p < 0.01, ○p < 0.05; Expression levels were normalized to the respective gene expression level detected in whole retinal extracts (gray shaded background). Anxa, annexin.
Fig. 5.
Functional verification of the efficient protection of Müller cells against oxidative stress. A, Exemplary protein network generated with proteins significantly enriched in Müller cells and with anti-oxidant activity revealed a significant accumulation of novel putative protectants against oxidative stress such as folate hydrolase 1 (FOLH1), paraoxonase 2 (PON2) and peroxiredoxin 6 (PRDX6) in Müller cells together with typical Müller cell markers (e.g. CD44) and already described protective Müller cell-specific enzymes such glutathione S-transferase T1 (GSTT1) and glutathione S-transferase mu 5 (GSTM5) in the Müller cell enriched fraction. Extrac., extracellular; memb., membrane; cyto., cytosol. B, Comparison of gene expression levels of selected antioxidant candidate genes detected by qRT-PCR performed on samples from whole retinal RNA extracts. C, Results from qRT-PCR experiments detecting transcript levels of selected genes with putative antioxidant activity in samples from whole retinal extracts (gray bar), enriched Müller cells (white bars) and the Müller cell-depleted neuronal fraction (black bars). Values represent the mean ± S.E. from four–five independent experiments. For each experiment, cells were pooled from four retinae originating from two animals. The values given in brackets below the gene symbol indicate the mean factor of enrichment of the respective protein as identified by the proteomic analysis. Significant difference between expression in Müller cell-enriched samples compared with Müller cell-depleted neuronal fractions: **p < 0.01, *p < 0.05; Significant difference between expression in Müller cell-enriched samples compared with whole retinal extracts: ••p < 0.01, •p < 0.05; Significant difference between expression in Müller cell-depleted samples compared with whole retinal extracts: ○○p < 0.01, ○p < 0.05. D, The subcellular localization of selected candidate genes in retinal sections was delineated by co-immunolabelings with the Müller cell marker glutamine synthetase (GS). PRDX6 is specifically expressed in Müller cells at presumably high levels and distributed throughout the whole cell body including their endfeet (open arrowheads) as well as their inner (arrows) and outer stem processes. Labeling for FOLH1 was less intense, but clearly confined to putative Müller cell structures such as their endfeet lining the inner retinal surface (open arrowheads) or their cell processes in the outer plexiform layer (OPL). Scale bars, 20 μm. GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; ONL, outer nuclear layer; PRS, photoreceptor segments; RPE, retinal pigment epithelium. E, Characterization of reactive oxygen species (ROS) formation in response to H2O2 application in isolated retinal cell subtypes using CM-H2DCFDA. Left, representative images were taken from recording CM-H2DCFDA fluorescence detected in bipolar cells (BP), Müller cells (MC) and photoreceptors (PR) before and after 15 min of H2O2 (100 μ
m
)exposure. Scale bars, 20 μm. Right, time course of alterations in the fluorescence intensity in the respective cell type (n = 10 each). Whiskers indicate S.E. values. Comparison of ROS levels in Müller cells versus photoreceptors: ••p < 0.01; •••p < 0.001. Comparison of ROS levels in Müller cells versus bipolar cells: ○○p < 0.01. F, In line with our expression data pointing to an excellent protection of Müller cells against oxidative stress, statistical analysis reveals the lowest levels of ROS formation in Müller cells that was significantly less pronounced compared with the response detected in bipolar cells and photoreceptor cells - the latter being most susceptible to H2O2-induced ROS formation. Numbers given in the bars indicate the numbers of measured cells. Values represent the mean ± S.E. Experiments were repeated for cell isolations from three different animals. ***p < 0.001.
Similar articles
- Glucocorticoid receptors in the retina, Müller glia and the formation of Müller glia-derived progenitors.
Gallina D, Zelinka C, Fischer AJ. Gallina D, et al. Development. 2014 Sep;141(17):3340-51. doi: 10.1242/dev.109835. Epub 2014 Aug 1. Development. 2014. PMID: 25085975 Free PMC article. - The agonistic TSPO ligand XBD173 attenuates the glial response thereby protecting inner retinal neurons in a murine model of retinal ischemia.
Mages K, Grassmann F, Jägle H, Rupprecht R, Weber BHF, Hauck SM, Grosche A. Mages K, et al. J Neuroinflammation. 2019 Feb 18;16(1):43. doi: 10.1186/s12974-019-1424-5. J Neuroinflammation. 2019. PMID: 30777091 Free PMC article. - Retinal microglia signaling affects Müller cell behavior in the zebrafish following laser injury induction.
Conedera FM, Pousa AMQ, Mercader N, Tschopp M, Enzmann V. Conedera FM, et al. Glia. 2019 Jun;67(6):1150-1166. doi: 10.1002/glia.23601. Epub 2019 Feb 22. Glia. 2019. PMID: 30794326 - Glia-neuron interactions in the mammalian retina.
Vecino E, Rodriguez FD, Ruzafa N, Pereiro X, Sharma SC. Vecino E, et al. Prog Retin Eye Res. 2016 Mar;51:1-40. doi: 10.1016/j.preteyeres.2015.06.003. Epub 2015 Jun 23. Prog Retin Eye Res. 2016. PMID: 26113209 Review. - Glia of the human retina.
Reichenbach A, Bringmann A. Reichenbach A, et al. Glia. 2020 Apr;68(4):768-796. doi: 10.1002/glia.23727. Epub 2019 Dec 3. Glia. 2020. PMID: 31793693 Review.
Cited by
- Mitochondrial transfer between BMSCs and Müller promotes mitochondrial fusion and suppresses gliosis in degenerative retina.
Huang X, Luodan A, Gao H, He J, Ge L, Cha Z, Gong H, Lin X, Li H, Tang Y, Jiang D, Fan X, Xu H. Huang X, et al. iScience. 2024 Jun 20;27(7):110309. doi: 10.1016/j.isci.2024.110309. eCollection 2024 Jul 19. iScience. 2024. PMID: 39055937 Free PMC article. - De novo peptide sequencing by deep learning.
Tran NH, Zhang X, Xin L, Shan B, Li M. Tran NH, et al. Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):8247-8252. doi: 10.1073/pnas.1705691114. Epub 2017 Jul 18. Proc Natl Acad Sci U S A. 2017. PMID: 28720701 Free PMC article. - Out-of-Field Hippocampus from Partial-Body Irradiated Mice Displays Changes in Multi-Omics Profile and Defects in Neurogenesis.
Pazzaglia S, Tanno B, Antonelli F, Giardullo P, Babini G, Subedi P, Azimzadeh O, Khan ZN, Oleksenko K, Metzger F, Toerne CV, Traynor D, Medipally D, Meade AD, Kadhim M, Lyng FM, Tapio S, Saran A, Mancuso M. Pazzaglia S, et al. Int J Mol Sci. 2021 Apr 20;22(8):4290. doi: 10.3390/ijms22084290. Int J Mol Sci. 2021. PMID: 33924260 Free PMC article. - Cross-talk between monocyte invasion and astrocyte proliferation regulates scarring in brain injury.
Frik J, Merl-Pham J, Plesnila N, Mattugini N, Kjell J, Kraska J, Gómez RM, Hauck SM, Sirko S, Götz M. Frik J, et al. EMBO Rep. 2018 May;19(5):e45294. doi: 10.15252/embr.201745294. Epub 2018 Apr 9. EMBO Rep. 2018. PMID: 29632244 Free PMC article. - SMAD7 deficiency stimulates Müller progenitor cell proliferation during the development of the mammalian retina.
Kugler M, Schlecht A, Fuchshofer R, Schmitt SI, Kleiter I, Aigner L, Tamm ER, Braunger BM. Kugler M, et al. Histochem Cell Biol. 2017 Jul;148(1):21-32. doi: 10.1007/s00418-017-1549-5. Epub 2017 Mar 3. Histochem Cell Biol. 2017. PMID: 28258388
References
- Shen W., Fruttiger M., Zhu L., Chung S. H., Barnett N. L., Kirk J. K., Lee S., Coorey N. J., Killingsworth M., Sherman L. S., and Gillies M. C. (2012) Conditional Mullercell ablation causes independent neuronal and vascular pathologies in a novel transgenic model. J. Neurosci. 32, 15715–15727 - PMC - PubMed
- Pannicke T., Frommherz I., Biedermann B., Wagner L., Sauer K., Ulbricht E., Hartig W., Krugel U., Ueberham U., Arendt T., Illes P., Bringmann A., Reichenbach A., and Grosche A. (2014) Differential effects of P2Y1 deletion on glial activation and survival of photoreceptors and amacrine cells in the ischemic mouse retina. Cell Death Disease 5, e1353. - PMC - PubMed
- Pannicke T., Iandiev I., Uckermann O., Biedermann B., Kutzera F., Wiedemann P., Wolburg H., Reichenbach A., and Bringmann A. (2004) A potassium channel-linked mechanism of glial cell swelling in the postischemic retina. Mol. Cell. Neurosci. 26, 493–502 - PubMed
- Wurm A., Pannicke T., Iandiev I., Wiedemann P., Reichenbach A., and Bringmann A. (2006) The developmental expression of K+ channels in retinal glial cells is associated with a decrease of osmotic cell swelling. Glia 54, 411–423 - PubMed
- Bringmann A., Iandiev I., Pannicke T., Wurm A., Hollborn M., Wiedemann P., Osborne N. N., and Reichenbach A. (2009) Cellular signaling and factors involved in Muller cell gliosis: neuroprotective and detrimental effects. Prog. Retin. Eye Re.s 28, 423–451 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases