Rap1 and its effector RIAM are required for lymphocyte trafficking - PubMed (original) (raw)
Rap1 and its effector RIAM are required for lymphocyte trafficking
Wenjuan Su et al. Blood. 2015.
Abstract
Regulation of integrins is critical for lymphocyte adhesion to endothelium and trafficking through secondary lymphoid organs. Inside-out signaling to integrins is mediated by the small GTPase Rap1. Two effectors of Rap1 regulate integrins, RapL and Rap1 interacting adaptor molecule (RIAM). Using mice conditionally deficient in both Rap1a and Rap1b and mice null for RIAM, we show that the Rap1/RIAM module is not required for T- or B-cell development but is essential for efficient adhesion to intercellular adhesion molecule (ICAM) 1 and vascular cell adhesion molecule (VCAM) 1 and for proper trafficking of lymphocytes to secondary lymphoid organs. Interestingly, in RIAM-deficient mice, whereas peripheral lymph nodes (pLNs) were depleted of both B and T cells and recirculating B cells were diminished in the bone barrow (BM), the spleen was hypercellular, albeit with a relative deficiency of marginal zone B cells. The abnormality in lymphocyte trafficking was accompanied by defective humoral immunity to T-cell-dependent antigens. Platelet function was intact in RIAM-deficient animals. These in vivo results confirm a role for RIAM in the regulation of some, but not all, leukocyte integrins and suggest that RIAM-regulated integrin activation is required for trafficking of lymphocytes from blood into pLNs and BM, where relatively high shear forces exist in high endothelial venules and sinusoids, respectively.
© 2015 by The American Society of Hematology.
Figures
Figure 1
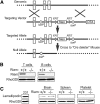
Rap1-deficient T cells develop normally but are defective in adhesion to ICAM-1 and homing to peripheral lymph nodes. (A) Immunoblot for Rap1 (antibody detects both Rap1a and Rap1b) of lysates of T cells, thymocytes, and splenocytes from mice of the indicated genotypes. β-tubulin serves as a loading control. (B) Percentages of thymocytes of animals with the indicated genotypes that were characterized by flow cytometry as CD4+, CD8+, double-positive, or double-negative. (C) Percentage of resting or stimulated (anti-CD3 or phorbol myristate acetate) splenic T cells from animals with the indicated genotypes adherent to ICAM-1 coated wells after washing. (D) Absolute number of CD3+ cells in thymus, 1 mL blood, spleen, and pLNs of animals with the indicated genotypes. (B, C, and D) Results plotted are mean ± SD; n = 4; **P < .01, ***P < .001.
Figure 2
Generation of RIAM−/− mice. (A) Schematic representation of RIAM targeting. The RIAM locus of murine ES cells was targeted by homologous recombination to generate an allele with loxP sites flanking exons 3 and 4. The resulting RIAM conditional mice were crossed with a Cre-deleter strain to remove exons 3 and 4, along with the Neo cassette, and thereby generate RIAM+/− mice that were crossed to produce RIAM−/− mice. (B) Immunmoblot for RIAM and RhoGDI of lysates of T and B cells isolated from spleens of RIAM+/+ and RIAM−/− mice. (C) Immunoblot for lamellipodin and RhoGDI of lysates of of MDA-MB-231 human breast cancer cells, as well as the extracts of brain, spleen, and platelets from mice with the indicated genotype.
Figure 3
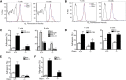
RIAM-deficient lymphocytes develop normally, but the MZ B cell population is reduced in RIAM-deficient mice. (A) RIAM+/+ and RIAM−/− thymocytes were stained with anti-CD4 and anti-CD8 Abs and analyzed by flow cytometry to reveal double-positive (DP), double-negative (DN), and single-positive (CD4+ or CD8+) cells. (B) Percentages of thymocyte subsets of animals with the indicated genotypes. Plotted are means ± SD; n = 4. (C) BM lymphocytes from RIAM+/+ or RIAM−/− mice were stained with anti-B220 and anti-IgM Abs (left) and analyzed by flow cytometry, depicting pre-pro (P; B220low, IgMlow), immature (I; B220high, IgMlow), and mature (M; B220high, IgMhigh) B-cell subsets or stained with anti-IgM and anti-IgD Abs (right) to quantify recirculating mature B cells. (D) Absolute numbers of B220+ B cells of the indicated subsets within equal volumes of BM suspensions; n = 4. (E, G) Splenocytes derived from RIAM+/+ and RIAM−/− mice were characterized by flow cytometry as total B cells or FO (CD23high, CD21low), or MZ (CD23low, CD21high) B cells (E) or T1 (IgMhigh, CD23low), T2 (IgMhigh, CD23high), or T3 (IgMlow, CD23high) transitional B cells (G). (F, H) Absolute numbers of the B cells characterized in E and G. Horizontal lines indicate mean ± SD of the individual animals plotted, *P < .05.
Figure 4
RIAM−/− lymphocytes are defective in integrin-dependent adhesion and migration. (A-B) Expression of LFA-1 (αL; A) and VLA-4 (α4; B) in spleen-derived T and B cells from RIAM+/+ or RIAM−/− mice was determined by flow cytometry. (C-D) Percentage of T (left) or B (right) cells (resting or stimulated as indicated) from spleens of RIAM+/+ or RIAM−/− mice adherent to ICAM-1 (C) or VCAM-1 (D) coated wells after washing. (E) T-cell adhesion to ICAM-1 ±SDF-1 measured in a flow chamber. (F) B-cell migration measured as percentage of cells that cross the membrane of a modified Boyden chamber toward media ± SDF-1. Plots show mean ± SD; n = 4 (C-D) or n = 3 (E-F). *P < .05, **P < .01, ***P < .001.
Figure 5
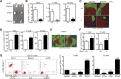
RIAM-deficient lymphocytes traffic efficiently to spleen but not pLNs or BM. (A) Brachial, axillary, and inguinal lymph nodes removed from RIAM+/+ or RIAM−/−mice. Scale bar represents 5 mm. (B, D) Numbers of T and B cells recovered from pLNs (B) or spleens (D) of RIAM+/+ or RIAM−/−mice. Plotted are mean ± SD; n = 7; *P < .05, **P < .01. (C, E) Thin sections of inguinal and brachial pLNs (C) or spleens (E) from RIAM+/+ or RIAM−/− mice stained for B cells (B220), T cells (CD3), and stromal cells (F480). Bar represents 100 μm. (F) Numbers of T and B cells in the peripheral blood of RIAM+/+ or RIAM−/−mice. Plotted are mean ± SD; n = 4. (G) Numbers of red (CMTMR stained RIAM+/+) or green (carboxyfluorescein diacetate succinimidyl ester stained RIAM−/−) CD3+ T cells and CD19+ B cells detected by flow cytometry in the spleen, pLNs, BM, and blood of RIAM+/+ recipients 3 hours after intravenous injection of equal numbers of stained, red blood cell-depleted splenocytes. On the left is a representative cytofluorimetric scatter plot of CD19+ cells, and on the right are cumulative data plotted as the mean ± SD of the ratios of green:red cells; n = 9; ***P < .001, ****P < .0001.
Figure 6
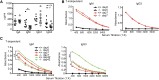
RIAM-deficient mice are defective in T-cell-dependent humoral immunity. (A) IgA, IgM, IgG1, and IgG3 levels in serum of naive RIAM+/+ (filled circles) and RIAM−/− (open squares) mice. Data from individual animals are plotted along the Y-axis with horizontal lines indicating mean ± SD. The difference in IgM levels was statistically significant (P < .05). (B-C) Anti-TNP IgM and IgG3 levels in serum of RIAM+/+ and RIAM−/− mice measured by titration and ELISA, 7 (B-C) and 42 (C) days after intraperitoneal injection with the T-cell-independent immunogen TNP-Ficoll (B) or the T-cell-dependent immunogen TNP-KLH (C). A boost of TNP-KLH was given at day 35 (C). Titers shown are representative of 4 mice of each genotype immunized with each antigen. Anti-TNP IgM and IgG3 levels in mice immunized with TNP-KLH differed significantly at day 7 and 42 (* P < .05, ** P < .01, 2-way ANOVA).
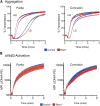
Figure 7
Intact platelet function in RIAM−/− mice. (A) Light transmission aggregometry of washed platelets obtained from heparinized whole blood of control or RIAM−/− mice after stimulation with low-dose (LD) or high-dose (HD) Par4p (50 or 100 µM) or convulxin (75 or 150 ng/mL). Four pairs of mice were studied, and representative curves are shown. (B) Real-time activation of the αIIbβ3 integrin after stimulation with Par4p or convulxin was measured in washed platelets using JON/A-PE. Combined data from 4 pairs of mice are shown.
Comment in
- The Rap1-RIAM pathway prefers β2 integrins.
Calderwood DA. Calderwood DA. Blood. 2015 Dec 17;126(25):2658-9. doi: 10.1182/blood-2015-09-668962. Blood. 2015. PMID: 26679542 Free PMC article.
Similar articles
- Structural, biochemical, and functional properties of the Rap1-Interacting Adaptor Molecule (RIAM).
Sari-Ak D, Torres-Gomez A, Yazicioglu YF, Christofides A, Patsoukis N, Lafuente EM, Boussiotis VA. Sari-Ak D, et al. Biomed J. 2022 Apr;45(2):289-298. doi: 10.1016/j.bj.2021.09.005. Epub 2021 Oct 1. Biomed J. 2022. PMID: 34601137 Free PMC article. Review. - Rap1-interacting adapter molecule (RIAM) associates with the plasma membrane via a proximity detector.
Wynne JP, Wu J, Su W, Mor A, Patsoukis N, Boussiotis VA, Hubbard SR, Philips MR. Wynne JP, et al. J Cell Biol. 2012 Oct 15;199(2):317-30. doi: 10.1083/jcb.201201157. Epub 2012 Oct 8. J Cell Biol. 2012. PMID: 23045549 Free PMC article. - RIAM links the ADAP/SKAP-55 signaling module to Rap1, facilitating T-cell-receptor-mediated integrin activation.
Ménasché G, Kliche S, Chen EJ, Stradal TE, Schraven B, Koretzky G. Ménasché G, et al. Mol Cell Biol. 2007 Jun;27(11):4070-81. doi: 10.1128/MCB.02011-06. Epub 2007 Apr 2. Mol Cell Biol. 2007. PMID: 17403904 Free PMC article. - HPK1 competes with ADAP for SLP-76 binding and via Rap1 negatively affects T-cell adhesion.
Patzak IM, Königsberger S, Suzuki A, Mak TW, Kiefer F. Patzak IM, et al. Eur J Immunol. 2010 Nov;40(11):3220-5. doi: 10.1002/eji.201040313. Eur J Immunol. 2010. PMID: 20957749 - The adaptor molecule RIAM integrates signaling events critical for integrin-mediated control of immune function and cancer progression.
Patsoukis N, Bardhan K, Weaver JD, Sari D, Torres-Gomez A, Li L, Strauss L, Lafuente EM, Boussiotis VA. Patsoukis N, et al. Sci Signal. 2017 Aug 22;10(493):eaam8298. doi: 10.1126/scisignal.aam8298. Sci Signal. 2017. PMID: 28831022 Review.
Cited by
- Direct Rap1/Talin1 interaction regulates platelet and neutrophil integrin activity in mice.
Bromberger T, Klapproth S, Rohwedder I, Zhu L, Mittmann L, Reichel CA, Sperandio M, Qin J, Moser M. Bromberger T, et al. Blood. 2018 Dec 27;132(26):2754-2762. doi: 10.1182/blood-2018-04-846766. Epub 2018 Nov 15. Blood. 2018. PMID: 30442677 Free PMC article. - SKAP2-A Molecule at the Crossroads for Integrin Signalling and Immune Cell Migration and Function.
Wilmink M, Spalinger MR. Wilmink M, et al. Biomedicines. 2023 Oct 14;11(10):2788. doi: 10.3390/biomedicines11102788. Biomedicines. 2023. PMID: 37893161 Free PMC article. Review. - Structural, biochemical, and functional properties of the Rap1-Interacting Adaptor Molecule (RIAM).
Sari-Ak D, Torres-Gomez A, Yazicioglu YF, Christofides A, Patsoukis N, Lafuente EM, Boussiotis VA. Sari-Ak D, et al. Biomed J. 2022 Apr;45(2):289-298. doi: 10.1016/j.bj.2021.09.005. Epub 2021 Oct 1. Biomed J. 2022. PMID: 34601137 Free PMC article. Review. - LFA1 Activation: Insights from a Single-Molecule Approach.
Kondo N, Ueda Y, Kinashi T. Kondo N, et al. Cells. 2022 May 26;11(11):1751. doi: 10.3390/cells11111751. Cells. 2022. PMID: 35681446 Free PMC article. Review. - Talin‑1 interaction network in cellular mechanotransduction (Review).
Zhao Y, Lykov N, Tzeng C. Zhao Y, et al. Int J Mol Med. 2022 May;49(5):60. doi: 10.3892/ijmm.2022.5116. Epub 2022 Mar 10. Int J Mol Med. 2022. PMID: 35266014 Free PMC article. Review.
References
- Mor A, Dustin ML, Philips MR. Small GTPases and LFA-1 reciprocally modulate adhesion and signaling. Immunol Rev. 2007;218:114–125. - PubMed
- Katagiri K, Maeda A, Shimonaka M, Kinashi T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat Immunol. 2003;4(8):741–748. - PubMed
- Lafuente EM, van Puijenbroek AA, Krause M, et al. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell. 2004;7(4):585–595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01-HL120846/HL/NHLBI NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- T32 HL007151/HL/NHLBI NIH HHS/United States
- R01 GM055279/GM/NIGMS NIH HHS/United States
- R01 HL121650/HL/NHLBI NIH HHS/United States
- GM58801/GM/NIGMS NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- T32 GM007308/GM/NIGMS NIH HHS/United States
- P30-CA016087/CA/NCI NIH HHS/United States
- P30-CA14051/CA/NCI NIH HHS/United States
- U54 CA112967/CA/NCI NIH HHS/United States
- GM055279/GM/NIGMS NIH HHS/United States
- R01 GM058801/GM/NIGMS NIH HHS/United States
- P01 HL120846/HL/NHLBI NIH HHS/United States
- U54-CA112967/CA/NCI NIH HHS/United States
- T32-GM007308/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous