Architecture of the Human and Yeast General Transcription and DNA Repair Factor TFIIH - PubMed (original) (raw)
. 2015 Sep 3;59(5):794-806.
doi: 10.1016/j.molcel.2015.07.016.
Peter Cimermancic 2, Shruthi Viswanath 2, Christopher C Ebmeier 3, Bong Kim 1, Marine Dehecq 4, Vishnu Raman 3, Charles H Greenberg 2, Riccardo Pellarin 2, Andrej Sali 2, Dylan J Taatjes 3, Steven Hahn 5, Jeff Ranish 6
Affiliations
- PMID: 26340423
- PMCID: PMC4560838
- DOI: 10.1016/j.molcel.2015.07.016
Architecture of the Human and Yeast General Transcription and DNA Repair Factor TFIIH
Jie Luo et al. Mol Cell. 2015.
Abstract
TFIIH is essential for both RNA polymerase II transcription and DNA repair, and mutations in TFIIH can result in human disease. Here, we determine the molecular architecture of human and yeast TFIIH by an integrative approach using chemical crosslinking/mass spectrometry (CXMS) data, biochemical analyses, and previously published electron microscopy maps. We identified four new conserved "topological regions" that function as hubs for TFIIH assembly and more than 35 conserved topological features within TFIIH, illuminating a network of interactions involved in TFIIH assembly and regulation of its activities. We show that one of these conserved regions, the p62/Tfb1 Anchor region, directly interacts with the DNA helicase subunit XPD/Rad3 in native TFIIH and is required for the integrity and function of TFIIH. We also reveal the structural basis for defects in patients with xeroderma pigmentosum and trichothiodystrophy, with mutations found at the interface between the p62 Anchor region and the XPD subunit.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
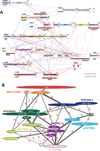
Figure 1. Purification and crosslinking maps of human and yeast TFIIH
A) Silver stained gel of purified human TFIIH. B) Map of the identified inter-protein (red lines) and intra-protein (blue lines) crosslinks for human TFIIH. C) Silver stained gel of purified yeast TFIIH. D) Inter- and intra-protein crosslink map for yeast TFIIH as in (B). Red and green dots indicate the positions of lysine residues. Red dots indicate that the lysine residue was identified in a crosslink.
Figure 2. Molecular architecture of human and yeast TFIIH
A) Localization density map of the human core TFIIH subunits (top) and the best scoring model (bottom). The EM density map of human TFIIH used in this study is shown in grey mesh. B) Domain decomposition of human core TFIIH. C) Localization density map of the yeast TFIIH subunits (top) and the best scoring model (bottom). The EM density map of yeast TFIIH used in this study is shown in grey mesh. D) Domain decomposition of yeast TFIIH. The human and yeast models are superposed on the XPD/Rad3 subunit. The different shapes between the two models are due to differences in the cryo-EM density maps and may not represent actual structural differences between the two species, but are possibly due to artifacts due to different sample preparations and data collection as well as processing.
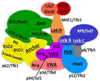
Figure 3. Summary of CXMS results for human and yeast TFIIH
A) Map of the identified inter-protein crosslinks for human (red lines) and yeast TFIIH (blue lines). Aligned sequences of human (bottom) and yeast (top) subunits are shown. Known and predicted structural domains are indicated as well as the positions of conserved topological regions identified in this study (orange bars). B) Summary of identified domain-domain crosslinks for human and yeast TFIIH. The thickness of the line is proportional to the number of crosslinks identified for each domain-domain linkage. The number of crosslinks supporting each domain-domain linkage is provided.
Figure 4. Crosslinks involving p62/Tfb1, XPD/Rad3, p44/Ssl1, and p34/Tfb4
Identified inter- and intra-protein crosslinks for human (red lines) and yeast (blue lines) homologs are mapped onto their aligned sequences. Known and predicted structural domains are indicated as well as the position of the p62/Tfb1Anchor region (red bar).
Figure 5. The p62/Tfb1 Anchor region is essential for the structural integrity of TFIIH
A) Schematic of the Tfb1 serial deletions analyzed in this study along with results of growth and UV sensitivity assays. Red bar indicates lethal phenotype. B) Results of IP-western analyses of Tfb1 domain deletions. The results are normalized to IP levels in the wild type Tfb1 strain. Error bars represent SEM for n = 3. C) Co-immunoprecipitation analysis with flag-tagged WT p62 (lane 2) or Anchor region deletions 292–328 (lane 3) or 328–432 (lane 4) in HeLa cells. CTRL: untransfected cells (lane 1). Note human p62 residues 292–328 and 328–432 align to yeast Tfb1 residues 359–397 and 397–505, respectively.
Figure 6. Schematic model of TFIIH subunit organization highlighting conserved topological features
The conserved topological features identified in this study are underlined. Subunit coloring scheme is the same as in Figure 2.
Figure 7. Mapping disease-associated mutations in XPD onto the human TFIIH models
Average placement of XPD without its C-terminal domain (precision of 8.5 Å) in the human TFIIH models is shown on the left. Disease-associated mutations in XPD are shown as red spheres. The location of the subunits in the context of core TFIIH is indicated by the grey rectangle on the right.
Similar articles
- Function of Conserved Topological Regions within the Saccharomyces cerevisiae Basal Transcription Factor TFIIH.
Warfield L, Luo J, Ranish J, Hahn S. Warfield L, et al. Mol Cell Biol. 2016 Sep 12;36(19):2464-75. doi: 10.1128/MCB.00182-16. Print 2016 Oct 1. Mol Cell Biol. 2016. PMID: 27381459 Free PMC article. - XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase.
Fuss JO, Tainer JA. Fuss JO, et al. DNA Repair (Amst). 2011 Jul 15;10(7):697-713. doi: 10.1016/j.dnarep.2011.04.028. Epub 2011 May 14. DNA Repair (Amst). 2011. PMID: 21571596 Free PMC article. Review. - The rem mutations in the ATP-binding groove of the Rad3/XPD helicase lead to Xeroderma pigmentosum-Cockayne syndrome-like phenotypes.
Herrera-Moyano E, Moriel-Carretero M, Montelone BA, Aguilera A. Herrera-Moyano E, et al. PLoS Genet. 2014 Dec 11;10(12):e1004859. doi: 10.1371/journal.pgen.1004859. eCollection 2014 Dec. PLoS Genet. 2014. PMID: 25500814 Free PMC article. - Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair.
Coin F, Oksenych V, Egly JM. Coin F, et al. Mol Cell. 2007 Apr 27;26(2):245-56. doi: 10.1016/j.molcel.2007.03.009. Mol Cell. 2007. PMID: 17466626 - A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor.
Egly JM, Coin F. Egly JM, et al. DNA Repair (Amst). 2011 Jul 15;10(7):714-21. doi: 10.1016/j.dnarep.2011.04.021. Epub 2011 May 17. DNA Repair (Amst). 2011. PMID: 21592869 Review.
Cited by
- Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
van Eeuwen T, Shim Y, Kim HJ, Zhao T, Basu S, Garcia BA, Kaplan CD, Min JH, Murakami K. van Eeuwen T, et al. Nat Commun. 2021 Jun 7;12(1):3338. doi: 10.1038/s41467-021-23684-x. Nat Commun. 2021. PMID: 34099686 Free PMC article. - Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Yang C, Fujiwara R, Kim HJ, Basnet P, Zhu Y, Gorbea Colón JJ, Steimle S, Garcia BA, Kaplan CD, Murakami K. Yang C, et al. Mol Cell. 2022 Feb 3;82(3):660-676.e9. doi: 10.1016/j.molcel.2021.12.020. Epub 2022 Jan 19. Mol Cell. 2022. PMID: 35051353 Free PMC article. - Structure of an RNA polymerase II preinitiation complex.
Murakami K, Tsai KL, Kalisman N, Bushnell DA, Asturias FJ, Kornberg RD. Murakami K, et al. Proc Natl Acad Sci U S A. 2015 Nov 3;112(44):13543-8. doi: 10.1073/pnas.1518255112. Epub 2015 Oct 19. Proc Natl Acad Sci U S A. 2015. PMID: 26483468 Free PMC article. - An Open Data Format for Visualization and Analysis of Cross-Linked Mass Spectrometry Results.
Hoopmann MR, Mendoza L, Deutsch EW, Shteynberg D, Moritz RL. Hoopmann MR, et al. J Am Soc Mass Spectrom. 2016 Nov;27(11):1728-1734. doi: 10.1007/s13361-016-1435-8. Epub 2016 Jul 28. J Am Soc Mass Spectrom. 2016. PMID: 27469004 Free PMC article. - Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Yan C, Dodd T, He Y, Tainer JA, Tsutakawa SE, Ivanov I. Yan C, et al. Nat Struct Mol Biol. 2019 Jun;26(6):397-406. doi: 10.1038/s41594-019-0220-3. Epub 2019 May 20. Nat Struct Mol Biol. 2019. PMID: 31110295 Free PMC article.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. - PubMed
- Bedez F, Linard B, Brochet X, Ripp R, Thompson JD, Moras D, Lecompte O, Poch O. Functional insights into the core-TFIIH from a comparative survey. Genomics. 2013;101:178–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM053451/GM/NIGMS NIH HHS/United States
- P30 CA015704/CA/NCI NIH HHS/United States
- T32 GM008284/GM/NIGMS NIH HHS/United States
- T32 GM067547/GM/NIGMS NIH HHS/United States
- R21CA175849/CA/NCI NIH HHS/United States
- R01 GM053451/GM/NIGMS NIH HHS/United States
- R01 GM083960/GM/NIGMS NIH HHS/United States
- T32 EB009383/EB/NIBIB NIH HHS/United States
- P50 GM076547/GM/NIGMS NIH HHS/United States
- Howard Hughes Medical Institute/United States
- R21 CA175849/CA/NCI NIH HHS/United States
- P41 GM109824/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials