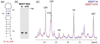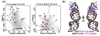N(6)-Methyladenosine Modification in a Long Noncoding RNA Hairpin Predisposes Its Conformation to Protein Binding - PubMed (original) (raw)
N(6)-Methyladenosine Modification in a Long Noncoding RNA Hairpin Predisposes Its Conformation to Protein Binding
Katherine I Zhou et al. J Mol Biol. 2016.
Abstract
N(6)-Methyladenosine (m(6)A) is a reversible and abundant internal modification of messenger RNA (mRNA) and long noncoding RNA (lncRNA) with roles in RNA processing, transport, and stability. Although m(6)A does not preclude Watson-Crick base pairing, the N(6)-methyl group alters the stability of RNA secondary structure. Since changes in RNA structure can affect diverse cellular processes, the influence of m(6)A on mRNA and lncRNA structure has the potential to be an important mechanism for m(6)A function in the cell. Indeed, an m(6)A site in the lncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) was recently shown to induce a local change in structure that increases the accessibility of a U5-tract for recognition and binding by heterogeneous nuclear ribonucleoprotein C (HNRNPC). This m(6)A-dependent regulation of protein binding through a change in RNA structure, termed "m(6)A-switch", affects transcriptome-wide mRNA abundance and alternative splicing. To further characterize this first example of an m(6)A-switch in a cellular RNA, we used nuclear magnetic resonance and Förster resonance energy transfer to demonstrate the effect of m(6)A on a 32-nucleotide RNA hairpin derived from the m(6)A-switch in MALAT1. The observed imino proton nuclear magnetic resonance resonances and Förster resonance energy transfer efficiencies suggest that m(6)A selectively destabilizes the portion of the hairpin stem where the U5-tract is located, increasing the solvent accessibility of the neighboring bases while maintaining the overall hairpin structure. The m(6)A-modified hairpin has a predisposed conformation that resembles the hairpin conformation in the RNA-HNRNPC complex more closely than the unmodified hairpin. The m(6)A-induced structural changes in the MALAT1 hairpin can serve as a model for a large family of m(6)A-switches that mediate the influence of m(6)A on cellular processes.
Keywords: FRET; MALAT1 lncRNA; N(6)-methyladenosine (m(6)A); NMR; RNA structural modeling.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
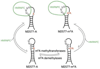
Fig. 1. The m6A-switch model
The human lncRNA MALAT1 is reversibly methylated at position A2577. The protein HNRNPC binds to the U5-tract in this hairpin from MALAT1, with an ~8-fold higher affinity for the methylated hairpin. One of the U’s in the HN NPC binding site pairs with the methylation site A2577. The presence of m6A weakens the base pair and increases the accessibility of the U-tract for protein binding.
Fig. 2. 1D NMR spectra show that the overall structure of the hairpin is maintained
(a) Secondary structure of the 32-nt M2577-A oligo from nucleotides 2556–2587 of MALAT1. The m6A modification site (A22 in the oligo, or A2577 in MALAT1) is denoted with a red “X.” The figure was made using Visualization Applet for RNA (VARNA) [38]. (b) 15% native PAGE of the unmethylated (M2577-A) and methylated (M2577-m6A) hairpins in 25 mM Tris acetate pH 7.4, 2.5 mM magnesium acetate. (c) Superimposed imino regions of the 1D 1H NMR spectra of M2577-A (blue) and M2577-m6A (red). Watergate solvent suppression 1D 1H NMR spectra were measured under the conditions 1.12 mM RNA,10 mM Na2HPO4 pH 7.4, 2.5 mM MgCl2, 90% H2O / 10% D2O (v/v), 20 °C.
Fig. 3. 2D NOESY spectra show that the upper stem is more dynamic in the methylated than in the unmethylated M2577 hairpin
(a) Superimposed imino regions of the 2D 1H NOESY NMR spectra of M2577-A (blue) and M2577-m6A (red) in 10% D2O. The spectra of 0.47 mM RNA were measured at 4 °C with a 100 ms mixing time. (b) Superimposed imino regions of the 2D 1H NOESY NMR spectra of M2577-A and M2577-m6A in 10% D2O. The spectra of 1.12 mM RNA were measured at 20 °C with a 100 ms mixing time. (c) Separate and superimposed amino–imino regions of the 2D 1H NOESY NMR spectra of M2577-A and M2577-m6A in 10% D2O at 20 °C. (d) Superimposed H6/H8–H1′ regions of the 2D 1H NOESY NMR spectra of M2577-A and M2577-m6A in 100% D2O. The spectra of 0.78 mM RNA were measured at 20 °C with a 100 ms mixing time.
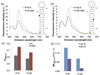
Fig. 4. FRET shows that the methylated and unmethylated MALAT1 hairpins have different conformations
(a) Fluorescence emission spectra of the FRET constructs Fl-8-A and Fl-8-m6A upon excitation at 490 nm. Cy3 (green) is conjugated to the 5′ phosphate, and fluorescein-dT (Fl) is incorporated at the indicated position (blue) in each oligo. Spectra were measured under the conditions 500 nM RNA, 10 mM Tris pH 7.5, 100 mM KCl, 2.5 mM MgCl2 at ambient temperature. Each spectrum is the average of 2–3 measurements. (b) Fluorescence emission spectra of Fl-26-A and Fl-26-m6A upon excitation at 490 nm. (c) Relative FRET efficiencies (EFRET,rel) of M2577-A and M2577-m6A, calculated as I563/(I563+I518), where I_x_ is the fluorescence emission intensity at x nm. FRET efficiencies are the mean of 6–8 measurements. Error bars represent ± one standard deviation. (d) Difference in the relative FRET efficiencies of M2577-A and M2577-m6A at ambient temperature (~20 °C) or at 90 °C.
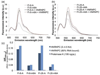
Fig. 5. FRET of RNPs containing the M2577-A and M2577-m6A hairpins
The RNPs show similar FRET, suggesting that the conformation of the RNA in the RNP is the same regardless of the presence of m6A. In addition, the methylated hairpins exhibit a smaller change in FRET upon HNRNPC binding. (a) Fluorescence emission spectra of 500 nM Fl-8-A and Fl-8-m6A with or without addition of HNRNPC at a concentration of 3–4.5·Kd (Kd = 722 nM for M2577-A, Kd = 93 nM for M2577-m6A) [19]. Spectra were measured under the conditions 500 nM RNA, 10 mM Tris pH 7.5, 100 mM KCl, 2.5 mM MgCl2 at ambient temperature at excitation wavelength 490 nm. (b) Fluorescence emission spectra of 500 nM Fl-26-A and Fl-26-m6A with or without addition of HNRNPC at a concentration of 3–4.5·Kd. (c) Change in the relative FRET efficiency (ΔEFRET,rel) of each hairpin (500 nM) upon addition of: 3–4.5·Kd HNRNPC (2.17 µM HNRNPC for M2577-A; 410 nM HNRNPC for M2577-m6A), HNRNPC such that [RNP]/[RNA]total = 80% (3.29 µM HNRNPC for M2577-A; 770 nM HNRNPC for M2577-m6A), or 106 ng/µL Proteinase K (equivalent to weight/volume concentration of 3.25 µM HNRNPC).
Fig. 6. Structural models for M2577-A and M2577-m6A based on FRET and NMR data
(a) Plot of 25 selected structures for M2577-A (gray) and M2577-m6A (dark red), in terms of κ2 and distance between fluorophores for Fl-8-A/m6A and Fl-26A/m6A. The structures were selected from an initial set of 9,999 tertiary structures for the M2577 hairpin [30] (c) Structural models of M2577-A (gray) and M2577-m6A (dark red), computed as the centroid of the 25 selected structures. The m6A modification site and the U5-tract are highlighted in shades of blue for the unmethylated MALAT1 hairpin, and in magenta for the methylated hairpin.
Similar articles
- N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions.
Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. Liu N, et al. Nature. 2015 Feb 26;518(7540):560-4. doi: 10.1038/nature14234. Nature. 2015. PMID: 25719671 Free PMC article. - Thermodynamic insights into N6-methyladenosine-modified ribonucleic acids and their interactions with the RNA recognition motif of heterogeneous nuclear ribonucleoprotein C.
Kumar A, Daripa P, Penumutchu S, Maiti S, Jain N. Kumar A, et al. Int J Biol Macromol. 2025 Jun;312:144210. doi: 10.1016/j.ijbiomac.2025.144210. Epub 2025 May 13. Int J Biol Macromol. 2025. PMID: 40373918 - Probing RNA Modification Status at Single-Nucleotide Resolution in Total RNA.
Liu N, Pan T. Liu N, et al. Methods Enzymol. 2015;560:149-59. doi: 10.1016/bs.mie.2015.03.005. Epub 2015 Jun 2. Methods Enzymol. 2015. PMID: 26253970 - N1-methyladenosine methylome in messenger RNA and non-coding RNA.
Xiong X, Li X, Yi C. Xiong X, et al. Curr Opin Chem Biol. 2018 Aug;45:179-186. doi: 10.1016/j.cbpa.2018.06.017. Epub 2018 Jul 9. Curr Opin Chem Biol. 2018. PMID: 30007213 Review. - Diverse molecular functions of m6A mRNA modification in cancer.
Han SH, Choe J. Han SH, et al. Exp Mol Med. 2020 May;52(5):738-749. doi: 10.1038/s12276-020-0432-y. Epub 2020 May 13. Exp Mol Med. 2020. PMID: 32404927 Free PMC article. Review.
Cited by
- Alteration of the N6-methyladenosine epitranscriptomic profile in synthetic phthalate-treated human induced pluripotent stem cell-derived endothelial cells.
Jousma J, Han Z, Yan G, Nukala SB, Kwon Y, Thi Le HH, Li Y, Ong SB, Lee WH, Ong SG. Jousma J, et al. Epigenomics. 2022 Oct;14(19):1139-1155. doi: 10.2217/epi-2022-0110. Epub 2022 Oct 31. Epigenomics. 2022. PMID: 36314267 Free PMC article. - The regulatory role of N6-methyladenosine RNA modification in gastric cancer: Molecular mechanisms and potential therapeutic targets.
Li G, Fu Q, Liu C, Peng Y, Gong J, Li S, Huang Y, Zhang H. Li G, et al. Front Oncol. 2022 Dec 6;12:1074307. doi: 10.3389/fonc.2022.1074307. eCollection 2022. Front Oncol. 2022. PMID: 36561529 Free PMC article. Review. - N⁶-methyladenosine (m⁶A): Revisiting the Old with Focus on New, an Arabidopsis thaliana Centered Review.
Bhat SS, Bielewicz D, Jarmolowski A, Szweykowska-Kulinska Z. Bhat SS, et al. Genes (Basel). 2018 Nov 30;9(12):596. doi: 10.3390/genes9120596. Genes (Basel). 2018. PMID: 30513695 Free PMC article. Review. - Function by Structure: Spotlights on Xist Long Non-coding RNA.
Pintacuda G, Young AN, Cerase A. Pintacuda G, et al. Front Mol Biosci. 2017 Dec 19;4:90. doi: 10.3389/fmolb.2017.00090. eCollection 2017. Front Mol Biosci. 2017. PMID: 29302591 Free PMC article. Review. - Programmable System of Cas13-Mediated RNA Modification and Its Biological and Biomedical Applications.
Tang T, Han Y, Wang Y, Huang H, Qian P. Tang T, et al. Front Cell Dev Biol. 2021 Jul 27;9:677587. doi: 10.3389/fcell.2021.677587. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34386490 Free PMC article. Review.
References
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DP1 GM105386/GM/NIGMS NIH HHS/United States
- K01 HG006699/HG/NHGRI NIH HHS/United States
- T32 GM007281/GM/NIGMS NIH HHS/United States
- K01HG006699/HG/NHGRI NIH HHS/United States
- NIGMS T32GM007281/PHS HHS/United States
- R01GM113194/GM/NIGMS NIH HHS/United States
- R01 GM113194/GM/NIGMS NIH HHS/United States
- DP1GM105386/DP/NCCDPHP CDC HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources