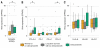Genomic correlates of response to CTLA-4 blockade in metastatic melanoma - PubMed (original) (raw)
. 2015 Oct 9;350(6257):207-211.
doi: 10.1126/science.aad0095. Epub 2015 Sep 10.
Eliezer M Van Allen # 1 2 3, Bastian Schilling # 4 5, Sachet A Shukla 1 2, Christian Blank 6, Lisa Zimmer 4 5, Antje Sucker 4 5, Uwe Hillen 4 5, Marnix H Geukes Foppen 6, Simone M Goldinger 7, Jochen Utikal 5 8 9, Jessica C Hassel 10, Benjamin Weide 11, Katharina C Kaehler 12, Carmen Loquai 13, Peter Mohr 14, Ralf Gutzmer 15, Reinhard Dummer 7, Stacey Gabriel 2, Catherine J Wu 1 2, Dirk Schadendorf 4 5, Levi A Garraway 1 2 3
Affiliations
- PMID: 26359337
- PMCID: PMC5054517
- DOI: 10.1126/science.aad0095
Genomic correlates of response to CTLA-4 blockade in metastatic melanoma
Eliezer M Van Allen et al. Science. 2015.
Erratum in
- Erratum for the Report "Genomic correlates of response to CTLA-4 blockade in metastatic melanoma" by E. M. Van Allen, D. Miao, B. Schilling, S. A. Shukla, C. Blank, L. Zimmer, A. Sucker, U. Hillen, M. H. Geukes Foppen, S. M. Goldinger, J. Utikal, J. C. Hassel, B. Weide, K. C. Kaehler, C. Loquai, P. Mohr, R. Gutzmer, R. Dummer, S. Gabriel, C. J. Wu, D. Schadendorf, L. A. Garraway.
[No authors listed] [No authors listed] Science. 2015 Nov 13;350(6262):aad8366. doi: 10.1126/science.aad8366. Science. 2015. PMID: 26564858 No abstract available. - Erratum for the Report "Genomic correlates of response to CTLA-4 blockade in metastatic melanoma" by E. M. Van Allen, D. Miao, B. Schilling, S. A. Shukla, C. Blank, L. Zimmer, A. Sucker, U. Hillen, M. H. Geukes Foppen, S. M. Goldinger, J. Utikal, J. C. Hassel, B. Weide, K. C. Kaehler, C. Loquai, P. Mohr, R. Gutzmer, R. Dummer, S. Gabriel, C. J. Wu, D. Schadendorf, L. A. Garraway.
[No authors listed] [No authors listed] Science. 2016 Apr 15;352(6283):aaf8264. doi: 10.1126/science.aaf8264. Science. 2016. PMID: 27081077 No abstract available.
Abstract
Monoclonal antibodies directed against cytotoxic T lymphocyte-associated antigen-4 (CTLA-4), such as ipilimumab, yield considerable clinical benefit for patients with metastatic melanoma by inhibiting immune checkpoint activity, but clinical predictors of response to these therapies remain incompletely characterized. To investigate the roles of tumor-specific neoantigens and alterations in the tumor microenvironment in the response to ipilimumab, we analyzed whole exomes from pretreatment melanoma tumor biopsies and matching germline tissue samples from 110 patients. For 40 of these patients, we also obtained and analyzed transcriptome data from the pretreatment tumor samples. Overall mutational load, neoantigen load, and expression of cytolytic markers in the immune microenvironment were significantly associated with clinical benefit. However, no recurrent neoantigen peptide sequences predicted responder patient populations. Thus, detailed integrated molecular characterization of large patient cohorts may be needed to identify robust determinants of response and resistance to immune checkpoint inhibitors.
Copyright © 2015, American Association for the Advancement of Science.
Figures
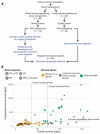
Fig. 1. Study design and clinical stratification
(A) Patients (n = 150) were identified for whole-exome sequencing of tumor and germline DNA. To be included in the original clinical cohort, patients had to have received ipilimumab monotherapy for metastatic cutaneous melanoma, have pretreatment germline and tumor samples available for sequencing, and have had overall survival for >14 days after initiation of ipilimumab therapy. Of these patients, 110 were eventually included in analysis after exclusions due to inadequate postsequencing quality control (n = 40) (18). Manual review of raw sequencing data was performed to exclude samples with evidence suggesting low purity, high contamination by ContEst (33), or discordant copy number quality control. Of the patients, 62, including 2 who failed DNA quality-control, had FFPE tumor samples available for transcriptome sequencing. After manual review for quality control following RNA sequencing, 42 samples were also analyzed for tumor microenvironment signatures, and 40 with matched WES were analyzed for neoantigen expression (14). (B) Patients were stratified into response groups based on RECIST criteria (21) (CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; MR, mixed response); duration of overall survival (OS); and duration of progression-free survival (PFS). All two-way comparisons were done comparing patients who achieved clinical benefit with ipilimumab (CR or PR by RECIST criteria or OS >1 year with SD by RECIST criteria) (n = 27) to those with minimal or no benefit from ipilimumab (PD by RECIST criteria or OS <1 year with SD by RECIST criteria) (n = 73). An additional cohort of patients who achieved long-term survival (OS >2 years) after ipilimumab treatment with early tumor progression (PFS <6 months) were considered separately (n = 10).
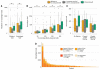
Fig. 2. Overall mutational load, overall neoantigen load, and expression-based neoantigen analysis as predictors of response to ipilimumab
(A) Elevated nonsynonymous mutational load and neoantigen load are associated with response to ipilimumab (P = 0.0076 and 0.027, respectively). An additional 20 points are not shown because of outlying high neoantigen loads in a subset of patients. (B) No trend in increased significance was observed when comparing the burden of higher-affinity neoantigens with respect to response to ipilimumab. Lower median inhibitory concentrations imply stronger HLA binding affinity on the x axis (P = 0.027 for affinity <500 nM; P = 0.034 for affinity <250 nM; P = 0.038 for affinity <100 nM; P = 0.042 for affinity <50 nM). An additional 34 points are not shown because of outlying high neoantigen loads in a subset of patients. (C) A sample size of 40 patients with complete DNA-sequencing, RNA-sequencing, and clinical annotation was insufficient to discern significant differences in neoantigen load or expressed neoantigen load among response cohorts, but a trend was observed for increased neoantigen load among patients with clinical benefit compared with those with no clinical benefit (P > 0.05 for all). (D) Patient-specific RNA-sequencing provides distinct information on tumor gene expression compared with TCGA melanoma data from a separate patient cohort. Although TCGA and RNA-seq data agree on the expression of the majority of neoantigens (n = 12,316) for 40 patients who had high-quality DNA- and RNA-sequencing data available for neoantigen and gene expression analysis, TCGA expression data overestimate the number of neoantigens expressed by 6320 in this patient cohort, and 166 neoantigens that are expressed by patient tumors would be missed by TCGA filtering alone. Additionally, a large proportion of neoantigens (n = 4349) are expressed at negligible levels in patient tumors. Asterisks (*) indicate P < 0.05.
Fig. 3. Immune microenvironment cytolytic and immune activity correlates with response to ipilimumab
(A) Patients who achieved clinical benefit from immune checkpoint blockade therapy had significantly higher levels of tumor cytolytic activity than those who had minimal or no benefit from ipilimumab (P = 0.039). (B) Patients who achieved clinical benefit from ipilimumab therapy had significantly higher levels of expression immune checkpoint receptors than those who did not (CTLA-4: P = 0.033, PD-L2: P = 0.041). One point is not shown because of an outlying high CTLA-4 expression value in a nonresponder patient (>50 reads per kilobase per million mapped reads). (C) Response to ipilimumab did not correlate with expression of or mutations in HLA alleles (P > 0.05 for all). Asterisks (*) indicate P < 0.05.
Comment in
- CANCER. The odds of immunotherapy success.
Gubin MM, Schreiber RD. Gubin MM, et al. Science. 2015 Oct 9;350(6257):158-9. doi: 10.1126/science.aad4140. Science. 2015. PMID: 26450194 No abstract available. - Tumor Microenvironment and Immunotherapy: The Whole Picture Is Better Than a Glimpse.
Church SE, Galon J. Church SE, et al. Immunity. 2015 Oct 20;43(4):631-3. doi: 10.1016/j.immuni.2015.10.004. Immunity. 2015. PMID: 26488814
Similar articles
- Genetic basis for clinical response to CTLA-4 blockade in melanoma.
Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Snyder A, et al. N Engl J Med. 2014 Dec 4;371(23):2189-2199. doi: 10.1056/NEJMoa1406498. Epub 2014 Nov 19. N Engl J Med. 2014. PMID: 25409260 Free PMC article. - Tumor Microenvironment and Immunotherapy: The Whole Picture Is Better Than a Glimpse.
Church SE, Galon J. Church SE, et al. Immunity. 2015 Oct 20;43(4):631-3. doi: 10.1016/j.immuni.2015.10.004. Immunity. 2015. PMID: 26488814 - Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer.
Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N, Hruban C, Guthrie VB, Rodgers K, Naidoo J, Kang H, Sharfman W, Georgiades C, Verde F, Illei P, Li QK, Gabrielson E, Brock MV, Zahnow CA, Baylin SB, Scharpf RB, Brahmer JR, Karchin R, Pardoll DM, Velculescu VE. Anagnostou V, et al. Cancer Discov. 2017 Mar;7(3):264-276. doi: 10.1158/2159-8290.CD-16-0828. Epub 2016 Dec 28. Cancer Discov. 2017. PMID: 28031159 Free PMC article. - CTLA-4 blockade increases antigen-specific CD8(+) T cells in prevaccinated patients with melanoma: three cases.
Yuan J, Ginsberg B, Page D, Li Y, Rasalan T, Gallardo HF, Xu Y, Adams S, Bhardwaj N, Busam K, Old LJ, Allison JP, Jungbluth A, Wolchok JD. Yuan J, et al. Cancer Immunol Immunother. 2011 Aug;60(8):1137-46. doi: 10.1007/s00262-011-1011-9. Epub 2011 Apr 5. Cancer Immunol Immunother. 2011. PMID: 21465316 Free PMC article. Review. - Targeting immune checkpoints in unresectable metastatic cutaneous melanoma: a systematic review and meta-analysis of anti-CTLA-4 and anti-PD-1 agents trials.
Yun S, Vincelette ND, Green MR, Wahner Hendrickson AE, Abraham I. Yun S, et al. Cancer Med. 2016 Jul;5(7):1481-91. doi: 10.1002/cam4.732. Epub 2016 May 11. Cancer Med. 2016. PMID: 27167347 Free PMC article. Review.
Cited by
- Construction of a novel lipid drop-mitochondria-associated genetic profile for predicting the survival and prognosis of lung adenocarcinoma.
Cai R, Lin H, Cheng Q, Mao Q, Zhang C, Tan Y. Cai R, et al. Discov Oncol. 2024 Nov 17;15(1):668. doi: 10.1007/s12672-024-01526-8. Discov Oncol. 2024. PMID: 39551861 Free PMC article. - E2F1-induced autocrine IL-6 inflammatory loop mediates cancer-immune crosstalk that predicts T cell phenotype switching and therapeutic responsiveness.
Spitschak A, Dhar P, Singh KP, Garduño RC, Gupta SK, Vera J, Musella L, Murr N, Stoll A, Pützer BM. Spitschak A, et al. Front Immunol. 2024 Oct 31;15:1470368. doi: 10.3389/fimmu.2024.1470368. eCollection 2024. Front Immunol. 2024. PMID: 39544930 Free PMC article. - The heterogeneity of NOTCH1 to tumor immune infiltration in pan-cancer.
Duan X, Wu R, Zhang M, Li K, Yu L, Sun H, Hao X, Wang C. Duan X, et al. Sci Rep. 2024 Nov 14;14(1):28071. doi: 10.1038/s41598-024-79883-1. Sci Rep. 2024. PMID: 39543218 Free PMC article. - Neoantigen architectures define immunogenicity and drive immune evasion of tumors with heterogenous neoantigen expression.
Roerden M, Castro AB, Cui Y, Harake N, Kim B, Dye J, Maiorino L, White FM, Irvine DJ, Litchfield K, Spranger S. Roerden M, et al. J Immunother Cancer. 2024 Nov 9;12(11):e010249. doi: 10.1136/jitc-2024-010249. J Immunother Cancer. 2024. PMID: 39521615 Free PMC article. - Recent Advances in Artificial Intelligence to Improve Immunotherapy and the Use of Digital Twins to Identify Prognosis of Patients with Solid Tumors.
D'Orsi L, Capasso B, Lamacchia G, Pizzichini P, Ferranti S, Liverani A, Fontana C, Panunzi S, De Gaetano A, Lo Presti E. D'Orsi L, et al. Int J Mol Sci. 2024 Oct 29;25(21):11588. doi: 10.3390/ijms252111588. Int J Mol Sci. 2024. PMID: 39519142 Free PMC article. Review.
References
- Robert C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–2526. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical