Metabolism and epigenetics - PubMed (original) (raw)
Review
Metabolism and epigenetics
Ryan Janke et al. Annu Rev Cell Dev Biol. 2015.
Abstract
Epigenetic mechanisms by which cells inherit information are, to a large extent, enabled by DNA methylation and posttranslational modifications of histone proteins. These modifications operate both to influence the structure of chromatin per se and to serve as recognition elements for proteins with motifs dedicated to binding particular modifications. Each of these modifications results from an enzyme that consumes one of several important metabolites during catalysis. Likewise, the removal of these marks often results in the consumption of a different metabolite. Therefore, these so-called epigenetic marks have the capacity to integrate the expression state of chromatin with the metabolic state of the cell. This review focuses on the central roles played by acetyl-CoA, S-adenosyl methionine, NAD(+), and a growing list of other acyl-CoA derivatives in epigenetic processes. We also review how metabolites that accumulate as a result of oncogenic mutations are thought to subvert the epigenetic program.
Keywords: S-adenosyl methionine; acetylation; chromatin modification; folate; methylation; oncometabolite.
Figures
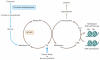
Figure 1
The one-carbon pathway (brown arrows) is important for the production of methionine and cysteine, as well as the SAM used in DNA and histone methylation reactions. Several inputs into the pathway replenish metabolites consumed during one-carbon metabolism. In cells expressing threonine dehydrogenase, threonine is oxidized to produce glycine, which is converted to methylene-THF (blue arrows). Folate obtained through diet or by de novo synthesis is converted into THF. Abbreviations: Me, methyl; MTHFR, methylenetetrahydrofolate reductase; SAH, S-adenosyl homocysteine; SAM, S-adenosyl methionine; THF, tetrahydrofolate.
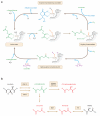
Figure 2
Potential roles of ascorbate and oncometabolites in regulating the activity of FeII- and α-ketoglutarate–dependent dioxygenases. (a) Following the oxidative decarboxylation of α-ketoglutarate, the dioxygenase can regain enzyme activity either by hydroxylating a substrate such as 5mC or by oxidizing ascorbate. (b) Mutations in IDH, SDH, and FH genes can result in high levels of oncometabolites (red) known to inhibit FeII- and α-ketoglutarate–dependent dioxygenases competitively with respect to α-ketoglutarate (green). Abbreviations: 5hmC, 5-hydroxymethylcytosine; 5mC, 5-methylcytosine; FH, fumarate hydratase; IDH, isocitrate dehydrogenase; SDH, succinate dehydrogenase.
Figure 3
Metabolic pathways used for synthesis of NAD+ in yeast and mammals. (a) The de novo NAD+ synthesis pathway converts tryptophan to quinolinate (blue box), which is then converted into NAMN, a precursor to NAD+. The NAD+ salvage pathway (light brown arrows) converts NAM, one of the by-products of sirtuin-mediated deacetylation, back into NAD+. Yeast converts NAM into NAD+ through the generation of NA. (b) Mammals are able to process NA in the same manner as yeast to generate NAD+. However, the mammalian salvage pathway does not generate NA; instead, NAM is converted into NMN and then directly back into NAD+. NAD+ is consumed during the deacetylation of histones catalyzed by sirtuins. Abbreviations: NA, nicotinic acid; NaAD, nicotinic acid adenine dinucleotide; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide; NAMN, nicotinic acid mononucleotide; NAMPT, nicotinamide phosphoribosyltransferase; NMN, nicotinamide mononucleotide; OAAR, _O-_acetyl ADP-ribose.
Similar articles
- Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression.
Donohoe DR, Bultman SJ. Donohoe DR, et al. J Cell Physiol. 2012 Sep;227(9):3169-77. doi: 10.1002/jcp.24054. J Cell Physiol. 2012. PMID: 22261928 Free PMC article. Review. - Linking epigenetics to lipid metabolism: focus on histone deacetylases.
Ferrari A, Fiorino E, Giudici M, Gilardi F, Galmozzi A, Mitro N, Cermenati G, Godio C, Caruso D, De Fabiani E, Crestani M. Ferrari A, et al. Mol Membr Biol. 2012 Nov;29(7):257-66. doi: 10.3109/09687688.2012.729094. Epub 2012 Oct 24. Mol Membr Biol. 2012. PMID: 23095054 Review. - Sink into the Epigenome: Histones as Repositories That Influence Cellular Metabolism.
Ye C, Tu BP. Ye C, et al. Trends Endocrinol Metab. 2018 Sep;29(9):626-637. doi: 10.1016/j.tem.2018.06.002. Epub 2018 Jul 11. Trends Endocrinol Metab. 2018. PMID: 30001904 Free PMC article. Review. - Compartmentalised acyl-CoA metabolism and roles in chromatin regulation.
Trefely S, Lovell CD, Snyder NW, Wellen KE. Trefely S, et al. Mol Metab. 2020 Aug;38:100941. doi: 10.1016/j.molmet.2020.01.005. Epub 2020 Feb 14. Mol Metab. 2020. PMID: 32199817 Free PMC article. Review. - Epigenetic Regulation: A New Frontier for Biomedical Engineers.
Chen Z, Li S, Subramaniam S, Shyy JY, Chien S. Chen Z, et al. Annu Rev Biomed Eng. 2017 Jun 21;19:195-219. doi: 10.1146/annurev-bioeng-071516-044720. Epub 2017 Mar 6. Annu Rev Biomed Eng. 2017. PMID: 28301736 Review.
Cited by
- Dri1 mediates heterochromatin assembly via RNAi and histone deacetylation.
Ban H, Sun W, Chen YH, Chen Y, Li F. Ban H, et al. Genetics. 2021 May 17;218(1):iyab032. doi: 10.1093/genetics/iyab032. Genetics. 2021. PMID: 33693625 Free PMC article. - Epigenetic Regulation of Ferroptosis in Central Nervous System Diseases.
Lan T, Sun TT, Wei C, Cheng T, Yang F, Zhang JN, Li Q. Lan T, et al. Mol Neurobiol. 2023 Jul;60(7):3584-3599. doi: 10.1007/s12035-023-03267-1. Epub 2023 Feb 27. Mol Neurobiol. 2023. PMID: 36847936 Review. - The Molecular Regulatory Mechanism in Multipotency and Differentiation of Wharton's Jelly Stem Cells.
Ma L, He X, Wu Q. Ma L, et al. Int J Mol Sci. 2023 Aug 18;24(16):12909. doi: 10.3390/ijms241612909. Int J Mol Sci. 2023. PMID: 37629090 Free PMC article. Review. - Demethylation of the Protein Phosphatase PP2A Promotes Demethylation of Histones to Enable Their Function as a Methyl Group Sink.
Ye C, Sutter BM, Wang Y, Kuang Z, Zhao X, Yu Y, Tu BP. Ye C, et al. Mol Cell. 2019 Mar 21;73(6):1115-1126.e6. doi: 10.1016/j.molcel.2019.01.012. Epub 2019 Feb 13. Mol Cell. 2019. PMID: 30772176 Free PMC article. - Nfe2l2 Regulates Metabolic Rewiring and Epigenetic Reprogramming in Mediating Cancer Protective Effect by Fucoxanthin.
Wang L, Wu R, Sargsyan D, Su S, Kuo HC, Li S, Chou P, Sarwar MS, Phadnis A, Wang Y, Su X, Kong AN. Wang L, et al. AAPS J. 2022 Jan 18;24(1):30. doi: 10.1208/s12248-022-00679-0. AAPS J. 2022. PMID: 35043283
References
- Adam J, Yang M, Soga T, Pollard PJ. Rare insights into cancer biology. Oncogene. 2014;33(20):2547–56. - PubMed
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, et al. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 2002;277(21):18881–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources