The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy - PubMed (original) (raw)
The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy
Jin-Mi Heo et al. Mol Cell. 2015.
Abstract
Damaged mitochondria are detrimental to cellular homeostasis. One mechanism for removal of damaged mitochondria involves the PINK1-PARKIN pathway, which poly-ubiquitylates damaged mitochondria to promote mitophagy. We report that assembly of ubiquitin chains on mitochondria triggers autophagy adaptor recruitment concomitantly with activation of the TBK1 kinase, which physically associates with OPTN, NDP52, and SQSTM1. TBK1 activation in HeLa cells requires OPTN and NDP52 and OPTN ubiquitin chain binding. In addition to the known role of S177 phosphorylation in OPTN on ATG8 recruitment, TBK1-dependent phosphorylation on S473 and S513 promotes ubiquitin chain binding in vitro as well as TBK1 activation, OPTN mitochondrial retention, and efficient mitophagy in vivo. These data reveal a self-reinforcing positive feedback mechanism that coordinates TBK1-dependent autophagy adaptor phosphorylation with the assembly of ubiquitin chains on mitochondria to facilitate efficient mitophagy, and mechanistically links genes mutated in Parkinson's disease and amyotrophic lateral sclerosis in a common selective autophagy pathway.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
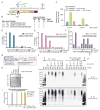
Figure 1. Activation of TBK1 upon mitochondrial depolarization requires PARKIN and PINK1
(A,B) HFT-MFN2−/− cells expressing an inducible MFN2 (WT or 8KR mutant)-IRES-PARKIN (WT or CS mutant) were depolarized (1h) and purified mitochondria (20μg) subjected to immunoblotting with MFN2 or TOMM20 antibodies (Panel A). In parallel experiments, cells were either left untreated, exposed to AO (1h) or transfected with Poly(I:C) (10μg/ml, 1.5h) and lysated subjected to immunoblotting with the indicated antibodies (Panel B). (C) Schematic of cell system used in this work. HeLa Flp-in T-Rex (HFT) cells harboring PARKINWT or catalytically inactive PARKINCS were created in the context of PINK1+/+ or PINK1−/− generated using gene-editing with a TALEN targeting PINK1. (D,E) HFT or HFT-PARKINWT cells were treated with AO (2h) to depolarize mitochondria or Poly(I:C) (2h) to activate the RIG-I/MDA5/MAVS pathway. Lysates were subjected to immunoblotting with the indicated antibodies (Panel D). Quantification of relative p-TBK1S172/TBK1 levels for biological triplicate experiments (Panel E). Error bars represent SEM from triplicate experiments. (F,G) HFT cells with the indicated PARKIN and PINK1 genotypes were depolarized with AO (1h) and lysates subjected to immunoblotting with the indicated antibodies (Panel F). Quantification of relative p-TBK1/TBK1 levels for biological triplicate experiments (Panel G). Error bars represent SEM from triplicate experiments. (H) U2OS cells stably expressing PARKINWT or SH-SY5Y cells were left untreated or depolarized with AO (1h) prior to immunoblotting of lysates. The p-TBK1S172/TBK1 ratios are indicated. Panels E and G analyzed by one-way ANOVA with Dunnett’s multiple comparisons test (n=3 biological replicates). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. n.s., not significant. See also Figure S1.
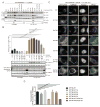
Figure 2. Recruitment of mitophagy adaptors OPTN NDP52, and SQSTM1, but not TAX1BP1, to depolarized mitochondria requires TBK1.<
br>(A–D) The indicated HFT cells stably expressing FLAG-HA-OPTN (Panel A) or FLAG-HA-NDP52 (Panel C) were left untreated or depolarized for 1h with AO in the presence or absence of the TBK1 inhibitor MRT (2μM, pre-treatment time 1h). Cells were imaged by confocal microscopy after staining with α-HA (green), α-TOMM20 (red), and Hoechst to detect DNA (blue) (scale bar, 20 microns). Normalized MOC values for co-localization of α-HA and α-TOMM20 were determined from >50 cells as described in Experimental Procedures. Panels B and D analyzed by one-way ANOVA with Dunnett’s multiple comparisons test. *p<0.05, **p<0.01, ****p<0.0001. n.s., not significant. Error bars represent SEM. (E,F) TBK1 is required for efficient recruitment of OPTN, NDP52, and SQSTM1, but not TAX1BP1 to depolarized mitochondria. Assays were performed as in Panel A in HFT-PARKINWT;TBK1−/− cells (scale bar, 20 microns). Panel F displays MOC values for SQSTM1 and TAX1BP1 co-localization with TOMM20 (error bars represent SEM). (G) Co-localization of p-TBK1S172 (green) with TOMM20-positive mitochondria (red) in depolarized HFT-PARKINWT cells was examined by immunofluorescence and confocal microscopy prior to image analysis to determine the MOC. Error bars represent SEM from triplicate experiments. (scale bar, 20 microns). (H) FLAG-HA-OPTN localizes to a subset of mitochondrial domains occupied by p-S65 UB in response to depolarization. HFT-PARKINWT cells stably expressing FLAG-HA-OPTN were depolarized for 75 min with AO and subjected to immunofluorescence with α-HA, α-p-S65 UB, or Hoechst to detect nuclei. Scale bar, 20 microns. (I) MOC for localization of HA-OPTN with p-S65 UB and for localization of p-S65 UB puncta with HA-OPTN puncta. Error bars are SEM of MOCs obtained from the indicated number of cell. (J) Localization of p-S65 UB to mitochondrial domains requires PARKIN. HFT cells lacking PARKIN were depolarized for 75 min with AO and subjected to immunofluorescence with α-TOMM20, α-p-S65 UB, or Hoechst to detect nuclei. Scale bar, 20 microns. See also Figure S2.
Figure 3. TBK1 activation by mitochondrial depolarization requires OPTN and NDP52 and the ability of OPTN to bind poly-UB
(A) Domain structure of OPTN showing the location of the poly-UB chain defective mutation (D474N) as well as 2 patient-based mutations in the UBAN motif. (B) TBK1 phosphorylation on S172 in response to mitochondrial depolarization is defective in HFT-PARKINWT;OPTN−/− cells reconstituted with an OPTND474N mutant that lacks poly-UB binding. The results of triplicate experiments for p-TBK1S172/TBK1 levels are shown in the histogram. Error bars represent SEM from triplicate experiments. (C) HFT PARKIN (WT or CS mutants) with the indicated genotypes for OPTN and NDP52 were left untreated or treated for 75 min with AO and protein extracts subjected to immunoblotting and analysis as described in panel B. Analyzed by one-way ANOVA with Dunnett’s multiple comparisons test (n=3 biological replicates). **p<0.01, ****p<0.0001.
Figure 4. Phosphorylation of OPTN, NDP52, and SQSTM1 in response to mitochondrial depolarization is dependent upon PINK1, PARKIN, and TBK1
(A,B) HFT-PARKINWT cells with or without PINK1 were depolarized with AO (75 min) in the presence or absence of the TBK1 inhibitor MRT (2μM, pre-treatment time 1h) and extracts subjected to SDS-PAGE or electrophoresis on Phos-tag gels prior to immunoblotting with the indicated antibodies. The position of depolarization dependent phosphorylated forms of mitophagy adaptors are indicated by arrows (Panel B). (C,D) As in panels A and B except that HFT cells expressing either PARKINWT or PARKINCS in the presence or absence of TBK1 were employed. See also Figure S3.
Figure 5. Quantitative proteomics identifies depolarization-dependent phosphorylation sites in OPTN that promote poly-UB binding.<
br>(A) Domain structure of OPTN showing the major sites of phosphorylation as well as patient-based mutations in the UBAN motif. (B) Identification of phosphorylation sites in OPTN upon mitochondrial depolarization by LC-MS2. HFT-PARKINWT cells expressing FLAG-HA-OPTN were depolarized (75 min) and the FLAG-HA-OPTN protein isolated by immunoprecipitation prior to trypsinization and analysis by LC-MS2. (C) Quantification of mitochondrial depolarization-dependent OPTN phosphorylation on S177 and S513 using TMT-based LC-MS3 proteomics. Error bars represent SEM from triplicate experiments. (D–F) Quantification of mitochondrial depolarization-dependent OPTN phosphorylation on S177, S473 and S513. OPTNS473 and S513 phosphorylation quantification from immune complexes was performed using AQUA proteomics (D–E) with heavy reference synthetic peptides (Table S1). Normalization for total OPTN levels was performed by also subjecting the same tryptic digest to TMT analysis (F) and quantifying all OPTN peptides identified. Analysis of OPTNS177 phosphorylation (F) was performed using TMT. N.D. - not determined due to mutation of a residue in the peptide of interest. (G,H) 6His-GST-OPTN and its site-specific phosphorylated forms (p-S177, p-S473, p-S513, and p-S473/p-S513) were expressed in bacteria using a tRNAp-Ser suppressor system (G) and purified to near heterogeneity, as seen by staining of SDS-PAGE gels with Coomassie Blue (H). (I) Quantification of phosphorylated recombinant 6His-GST-OPTN proteins using TMT-based proteomics. (J) The indicated unphosphorylated or p-S65 phosphorylated K48 or K63 UB(2–7) chains were incubated with the indicated phosphorylated forms of GST-OPTN, and after washing, bound proteins were released and subjected to SDS-PAGE and immunoblotting with α-UB. See also Figure S4.
Figure 6. Role of OPTN phosphorylation in TBK1 activation and OPTN recruitment to depolarized mitochondria
(A) Phosphorylation site mutants in OPTN do not affect association of TBK1 with OPTN. The indicated FLAG-HA-OPTN mutants in phosphorylation sites or in D474 required for UB binding were expressed in HFT-PARKIN (WT or CS mutants) cells engineered to lack OPTN using CRISPR-Cas9. Cells were either left untreated or treated with AO for 75 min and α-FLAG immunoprecipitates (OPTN) probed with antibodies against endogenous TBK1. Lysates were also immunoblotted to demonstrate loading. (B) Phosphorylation of S473 and S513 is required for efficient TBK1 activation in response to mitochondrial depolarization. The indicated GFP-OPTN mutants in phosphorylation sites were expressed in HFT-PARKIN (WT or CS mutants) cells engineered to lack OPTN using CRISPR-Cas9. Cells were either left untreated or treated with AO for 90 min lysates subjected to immunoblotting with the indicated antibodies. Analyzed by one-way ANOVA with Dunnett’s multiple comparisons test (n=3 biological replicates). **p<0.01, ****p<0.0001, n.s., not significant. (C,D) OPTNS473A/S513A is defective in retention on depolarized mitochondria. In Panel C, HFT-PARKINWT;OPTN−/− cells stably expressing the indicated GFP-OPTN mutants were depolarized with AO (90 min) and subjected to immunofluorescence using the indicated antibodies or direct observation of GFP-OPTN proteins. Scale bar, 20 microns. In Panel D, MOC values were obtained for >50 cells from triplicate experiments as described under Experimental Procedures. Analyzed by one-way ANOVA with Dunnett’s multiple comparisons test (n=3 biological replicates). ****p<0.0001, n.s., not significant. See also Figure S5.
Figure 7. A TBK1-OPTN-NDP52 network is required for efficient mitophagy and recruitment of LC3B to depolarized mitochondria
(A) HFT cells expressing the indicated PARKIN proteins and harboring the indicated TBK1, OPTN, and NDP52 genotypes were treated with AO and stained for α-DNA to detect mitochondria after 31h. Image analysis was used to determine the fraction of cells displaying unaggregated, aggregated, and cleared mitochondria as described (Ordureau et al., 2015a). (B) HFT-PARKINWT;TBK1−/− cells alone or reconstituted with the indicated TBK1 protein were subjected to AO treatment (30h) and mitochondria monitored and quantified as in panel A. (C) HFT-PARKINWT;OPTN−/−;NDP52−/− cells alone or reconstituted with the indicated FLAG-HA-OPTN proteins were subjected to AO treatment (30h) and mitochondria monitored and quantified as in panel A. (D) OPTN and NDP52-dependent recruitment of the ATG8 protein LC3B to damaged mitochondria. Cells were stained with α-LC3B to mark autophagosomes (green), α-TOMM20 to mark mitochondria, and Hoechst (blue) to mark nuclei and imaged by confocal microscopy (scale bar, 20 microns). The fraction of cells showing co-localization of LC3B and TOMM20 staining was determined by image analysis examining 24,455 cells (WT) and 17,343 cells (OPTN−/−;NDP52−/−). (D) Schematic of a positive feedback mechanism describing the role of UB chains, mitophagy adaptor phosphorylation, and TBK1 phosphorylation in ATG8 recruitment and mitophagy. In Step 1, PINK1 and PARKIN are activated and collaborate to generate UB chains on the MOM in Step 2, including chains with K63 linkages. In Step 3, mitophagy adaptors are recruited to various poly-UB chains, and in this context, TBK1 is recruited to mitochondria and activated by phosphorylation on S172 (Step 4) in a manner that depends primarily upon OPTN and NDP52 recruitment to poly-UB chains. The kinase required for this is unknown. In Step 5, activated TBK1 phosphorylates OPTN and NDP52. Steps 4 and 5 may be concerted. Phosphorylation of OPTN on S473 stabilizes its binding to various poly-UB chains while phosphorylation of S177 promotes association with LC3B (Step 6). Analyzed by one-way ANOVA with Dunnett’s multiple comparisons test (n=3 biological replicates). ***p<0.001, ****p<0.0001. n.s., not significant. See also Figure S6.
Comment in
- Better Safe than Sorry: Interlinked Feedback Loops for Robust Mitophagy.
Manford AG, Rape M. Manford AG, et al. Mol Cell. 2015 Oct 1;60(1):1-2. doi: 10.1016/j.molcel.2015.09.016. Mol Cell. 2015. PMID: 26431022
Similar articles
- Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy.
Moore AS, Holzbaur EL. Moore AS, et al. Proc Natl Acad Sci U S A. 2016 Jun 14;113(24):E3349-58. doi: 10.1073/pnas.1523810113. Epub 2016 May 31. Proc Natl Acad Sci U S A. 2016. PMID: 27247382 Free PMC article. - Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria.
Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, Youle RJ, Dikic I. Richter B, et al. Proc Natl Acad Sci U S A. 2016 Apr 12;113(15):4039-44. doi: 10.1073/pnas.1523926113. Epub 2016 Mar 30. Proc Natl Acad Sci U S A. 2016. PMID: 27035970 Free PMC article. - Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy.
Ordureau A, Heo JM, Duda DM, Paulo JA, Olszewski JL, Yanishevski D, Rinehart J, Schulman BA, Harper JW. Ordureau A, et al. Proc Natl Acad Sci U S A. 2015 May 26;112(21):6637-42. doi: 10.1073/pnas.1506593112. Epub 2015 May 12. Proc Natl Acad Sci U S A. 2015. PMID: 25969509 Free PMC article. - The three 'P's of mitophagy: PARKIN, PINK1, and post-translational modifications.
Durcan TM, Fon EA. Durcan TM, et al. Genes Dev. 2015 May 15;29(10):989-99. doi: 10.1101/gad.262758.115. Genes Dev. 2015. PMID: 25995186 Free PMC article. Review. - PINK1/Parkin-mediated mitophagy in mammalian cells.
Eiyama A, Okamoto K. Eiyama A, et al. Curr Opin Cell Biol. 2015 Apr;33:95-101. doi: 10.1016/j.ceb.2015.01.002. Epub 2015 Feb 17. Curr Opin Cell Biol. 2015. PMID: 25697963 Review.
Cited by
- DJ-1 is an essential downstream mediator in PINK1/parkin-dependent mitophagy.
Imberechts D, Kinnart I, Wauters F, Terbeek J, Manders L, Wierda K, Eggermont K, Madeiro RF, Sue C, Verfaillie C, Vandenberghe W. Imberechts D, et al. Brain. 2022 Dec 19;145(12):4368-4384. doi: 10.1093/brain/awac313. Brain. 2022. PMID: 36039535 Free PMC article. - Methods to Monitor Mitophagy and Mitochondrial Quality: Implications in Cancer, Neurodegeneration, and Cardiovascular Diseases.
Patergnani S, Bonora M, Bouhamida E, Danese A, Marchi S, Morciano G, Previati M, Pedriali G, Rimessi A, Anania G, Giorgi C, Pinton P. Patergnani S, et al. Methods Mol Biol. 2021;2310:113-159. doi: 10.1007/978-1-0716-1433-4_9. Methods Mol Biol. 2021. PMID: 34096002 - Electrostatic and steric effects underlie acetylation-induced changes in ubiquitin structure and function.
Kienle SM, Schneider T, Stuber K, Globisch C, Jansen J, Stengel F, Peter C, Marx A, Kovermann M, Scheffner M. Kienle SM, et al. Nat Commun. 2022 Sep 16;13(1):5435. doi: 10.1038/s41467-022-33087-1. Nat Commun. 2022. PMID: 36114200 Free PMC article. - Mechanistic insights into selective autophagy pathways: lessons from yeast.
Farré JC, Subramani S. Farré JC, et al. Nat Rev Mol Cell Biol. 2016 Sep;17(9):537-52. doi: 10.1038/nrm.2016.74. Epub 2016 Jul 6. Nat Rev Mol Cell Biol. 2016. PMID: 27381245 Free PMC article. Review. - The ubiquitin proteoform problem.
Deol KK, Strieter ER. Deol KK, et al. Curr Opin Chem Biol. 2021 Aug;63:95-104. doi: 10.1016/j.cbpa.2021.02.015. Epub 2021 Apr 1. Curr Opin Chem Biol. 2021. PMID: 33813043 Free PMC article. Review.
References
- Belzil VV, Daoud H, Desjarlais A, Bouchard JP, Dupre N, Camu W, Dion PA, Rouleau GA. Analysis of OPTN as a causative gene for amyotrophic lateral sclerosis. Neurobiol Aging. 2011;32:555 e513–554. - PubMed
- Clark K, Peggie M, Plater L, Sorcek RJ, Young ER, Madwed JB, Hough J, McIver EG, Cohen P. Novel cross-talk within the IKK family controls innate immunity. Biochem J. 2011;434:93–104. - PubMed
- Cunningham CN, Baughman JM, Phu L, Tea JS, Yu C, Coons M, Kirkpatrick DS, Bingol B, Corn JE. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat Cell Biol. 2015;17:160–169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous