Genetic Heterogeneity in Algerian Human Populations - PubMed (original) (raw)
Genetic Heterogeneity in Algerian Human Populations
Asmahan Bekada et al. PLoS One. 2015.
Abstract
The demographic history of human populations in North Africa has been characterized by complex processes of admixture and isolation that have modeled its current gene pool. Diverse genetic ancestral components with different origins (autochthonous, European, Middle Eastern, and sub-Saharan) and genetic heterogeneity in the region have been described. In this complex genetic landscape, Algeria, the largest country in Africa, has been poorly covered, with most of the studies using a single Algerian sample. In order to evaluate the genetic heterogeneity of Algeria, Y-chromosome, mtDNA and autosomal genome-wide makers have been analyzed in several Berber- and Arab-speaking groups. Our results show that the genetic heterogeneity found in Algeria is not correlated with geography or linguistics, challenging the idea of Berber groups being genetically isolated and Arab groups open to gene flow. In addition, we have found that external sources of gene flow into North Africa have been carried more often by females than males, while the North African autochthonous component is more frequent in paternally transmitted genome regions. Our results highlight the different demographic history revealed by different markers and urge to be cautious when deriving general conclusions from partial genomic information or from single samples as representatives of the total population of a region.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
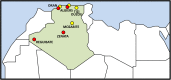
Fig 1. Geographic location of the Algerian samples genotyped in the present study (in red) and the samples obtained from the literature (in yellow).
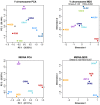
Fig 2. Bidimensional plots based on uniparental genomes.
PC analyses based on haplogroup data for Y-chromosome and mtDNA; and MDS analyses based on Y-STR haplotype data and on mtDNA sequence data. Abbreviations: ALG/ALG1: Algiers (this study), ALG2: Algiers (Y-chromosome; [10]), ORN1: Oran (present study), ORN2: Oran (Y-chromosome, [21]; mtDNA, [17]), RGB: Reguibate, ZNT: Zenata, MZB: Mozabite, TZO: Tizi Ouzou
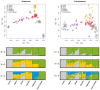
Fig 3. Plots for the analysis of genome-wide SNPs.
PC analysis (upper figures) based on autosomal data, and X-chromosome SNPs. ADMIXTURE proportions (bottom figures) at k = 2,3, and 4 based on autosomal data and X-chromosome SNPs. Algeria, stands for general Algerian sample [3]; Mozabite, stands for the Algerian Berber Mozabites [32]; and Zenata, stands for Algerian Berber Zenata (present study).
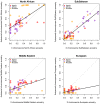
Fig 4. Correlation plots of the ancestry proportions at k = 4 in the ADMIXTURE analysis comparing autosomes and X-chromosome SNPs.
North African, sub-Saharan, Middle Eastern, and European ancestry proportions are shown in different plots. Solid black lines represent linear correlations between autosomal and X-chromosome components.
Similar articles
- Introducing the Algerian mitochondrial DNA and Y-chromosome profiles into the North African landscape.
Bekada A, Fregel R, Cabrera VM, Larruga JM, Pestano J, Benhamamouch S, González AM. Bekada A, et al. PLoS One. 2013;8(2):e56775. doi: 10.1371/journal.pone.0056775. Epub 2013 Feb 19. PLoS One. 2013. PMID: 23431392 Free PMC article. - Mitochondrial DNA heterogeneity in Tunisian Berbers.
Fadhlaoui-Zid K, Plaza S, Calafell F, Ben Amor M, Comas D, Bennamar El gaaied A. Fadhlaoui-Zid K, et al. Ann Hum Genet. 2004 May;68(Pt 3):222-33. doi: 10.1046/j.1529-8817.2004.00096.x. Ann Hum Genet. 2004. PMID: 15180702 - Female gene pools of Berber and Arab neighboring communities in central Tunisia: microstructure of mtDNA variation in North Africa.
Cherni L, Loueslati BY, Pereira L, Ennafaâ H, Amorim A, El Gaaied AB. Cherni L, et al. Hum Biol. 2005 Feb;77(1):61-70. doi: 10.1353/hub.2005.0028. Hum Biol. 2005. PMID: 16114817 - Mitochondrial DNA variation in Mauritania and Mali and their genetic relationship to other Western Africa populations.
González AM, Cabrera VM, Larruga JM, Tounkara A, Noumsi G, Thomas BN, Moulds JM. González AM, et al. Ann Hum Genet. 2006 Sep;70(Pt 5):631-57. doi: 10.1111/j.1469-1809.2006.00259.x. Ann Hum Genet. 2006. PMID: 16907709 - Population history of North Africa based on modern and ancient genomes.
Lucas-Sánchez M, Serradell JM, Comas D. Lucas-Sánchez M, et al. Hum Mol Genet. 2021 Apr 26;30(R1):R17-R23. doi: 10.1093/hmg/ddaa261. Hum Mol Genet. 2021. PMID: 33284971 Review.
Cited by
- CYP2C gene polymorphisms in North African populations.
Messaoudi M, Pakstis AJ, Boussetta S, Ben Ammar Elgaaied A, Kidd KK, Cherni L. Messaoudi M, et al. Mol Biol Rep. 2024 Nov 12;51(1):1145. doi: 10.1007/s11033-024-10093-8. Mol Biol Rep. 2024. PMID: 39532754 - Clinical and genetic spectrums of 413 North African families with inherited retinal dystrophies and optic neuropathies.
Bouzidi A, Charoute H, Charif M, Amalou G, Kandil M, Barakat A, Lenaers G. Bouzidi A, et al. Orphanet J Rare Dis. 2022 May 12;17(1):197. doi: 10.1186/s13023-022-02340-7. Orphanet J Rare Dis. 2022. PMID: 35551639 Free PMC article. Review. - Association of TERT, OGG1, and CHRNA5 Polymorphisms and the Predisposition to Lung Cancer in Eastern Algeria.
Mimouni A, Rouleau E, Saulnier P, Marouani A, Abdelali ML, Filali T, Beddar L, Lakehal A, Hireche A, Boudersa A, Aissaoui M, Ramtani H, Bouhedjar K, Abdellouche D, Oudjehih M, Boudokhane I, Abadi N, Satta D. Mimouni A, et al. Pulm Med. 2020 Mar 20;2020:7649038. doi: 10.1155/2020/7649038. eCollection 2020. Pulm Med. 2020. PMID: 32257438 Free PMC article. - Carriers of mitochondrial DNA macrohaplogroup R colonized Eurasia and Australasia from a southeast Asia core area.
Larruga JM, Marrero P, Abu-Amero KK, Golubenko MV, Cabrera VM. Larruga JM, et al. BMC Evol Biol. 2017 May 23;17(1):115. doi: 10.1186/s12862-017-0964-5. BMC Evol Biol. 2017. PMID: 28535779 Free PMC article. - Whole mitogenomes reveal that NW Africa has acted both as a source and a destination for multiple human movements.
Aizpurua-Iraola J, Abdeli A, Benhassine T, Calafell F, Comas D. Aizpurua-Iraola J, et al. Sci Rep. 2023 Jun 27;13(1):10395. doi: 10.1038/s41598-023-37549-4. Sci Rep. 2023. PMID: 37369751 Free PMC article.
References
- Camps G. Les civilisations préhistoriques de l’Afrique du Nord et du Sahara Paris: Doi; 1974. p. 374.
- Brett M, Fentress E. The Berbers. Oxford: Bl; 1996.
Publication types
MeSH terms
Substances
Grants and funding
This study was supported by the Ministerio de Economía y Competitividad grant CGL2013-44351-P and by Direcció General de Recerca, Generalitat de Catalunya grant 2014SGR866. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources