An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses - PubMed (original) (raw)
. 2015 Oct 8;163(2):381-93.
doi: 10.1016/j.cell.2015.08.061. Epub 2015 Sep 24.
Wendy Huang 1, Jason A Hall 1, Yi Yang 1, Alessandra Chen 2, Samuel J Gavzy 1, June-Yong Lee 1, Joshua W Ziel 1, Emily R Miraldi 3, Ana I Domingos 4, Richard Bonneau 5, Dan R Littman 6
Affiliations
- PMID: 26411290
- PMCID: PMC4621768
- DOI: 10.1016/j.cell.2015.08.061
An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses
Teruyuki Sano et al. Cell. 2015.
Erratum in
- Cell. 2016 Jan 14;164(1-2):324. Dosage error in article text
Abstract
RORγt(+) Th17 cells are important for mucosal defenses but also contribute to autoimmune disease. They accumulate in the intestine in response to microbiota and produce IL-17 cytokines. Segmented filamentous bacteria (SFB) are Th17-inducing commensals that potentiate autoimmunity in mice. RORγt(+) T cells were induced in mesenteric lymph nodes early after SFB colonization and distributed across different segments of the gastrointestinal tract. However, robust IL-17A production was restricted to the ileum, where SFB makes direct contact with the epithelium and induces serum amyloid A proteins 1 and 2 (SAA1/2), which promote local IL-17A expression in RORγt(+) T cells. We identified an SFB-dependent role of type 3 innate lymphoid cells (ILC3), which secreted IL-22 that induced epithelial SAA production in a Stat3-dependent manner. This highlights the critical role of tissue microenvironment in activating effector functions of committed Th17 cells, which may have important implications for how these cells contribute to inflammatory disease.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
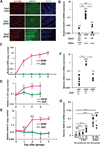
Figure 1. Site of SFB colonization correlates with maximal IL-17A induction in RORγt+ T cells
(A) Quantitative PCR (qPCR) analysis of SFB genomic 16S in duodenum, ileum and colon (proximal and distal segments of each) from SFB-colonized (SFBe; gavaged with SFB-rich fecal contents) or SFB-free (JAX; gavaged with SFB-free fecal contents) mice at 7 and 14 days post gavage. Copy number of SFB 16S was normalized by copy number of mouse genomic Il-23r gene. The experiment was repeated more than 3 times with similar results combined. Data are represented as mean with SD. (B) Number of RORγt+ CD4+ T cells in intestinal lamina propria (LP) or MLN of SFB-colonized (SFBe; n=7) or SFB-free (JAX; n=7) mice on 7 and 14 days post gavage. CD4+ T cells were isolated from the indicated tissues, and Foxp3 RORγt+ CD4+ T cells were quantified by FACS. (C) IL-17A (GFP+) production in Foxp3 RORγt+ CD4+ T cells from intestinal LPs and MLN in IL-17A reporter (Il17agfp/gfp) mice at 14 days post SFB colonization. IL-17A–producing Th17 cells were monitored by FACS after staining with anti-GFP antibody without re-stimulation. (D) The proportion of IL-17A producing Th17 cells among Foxp3− RORγt+ CD4+ T cells following SFB colonization. Each dot represents a single mouse, and the orange bar indicates average of relative gene expression of each group. Data were from at least two independent experiments (JAX; n=7. SFBe; n=9). N.S, not significant (P>0.05); *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001. See also Figure S1.
Figure 2. Role of epithelial SAA1/2 in ileal Th17 cell production of IL-17A
(A) Translating ribosome affinity purification (TRAP) assay for identification of the cell type expressing SAA1/2. Four individual lines of TRAP mice, conditionally expressing a GFP tagged ribosome L10 fusion protein in intestinal epithelial (Villin-Cre), T (CD4-Cre), myeloid (CD11c–Cre), or liver (Albumin-Cre) cells were generated. After two weeks of SFB colonization, ribosomes in small intestinal lysates collected from each line were enriched by GFP pull-down. Actively translating SAA1/2 mRNAs were quantified by qRT-PCR. Black and white bar indicates TRAP signal from SFB-free animals and light blue, dark blue, purple, and orange bars indicate TRAP signal from SFB positive animals. White bar indicates TRAP signal from tissue harvested from JAX animals at 6h following intraperitoneal administration of LPS. (B) Quantification of SAA1/2 mRNA in intestinal epithelial cells of duodenum, ileum and colon from SFBe (Day7; n=8, Day 14; n=3), pure SFB (n=4), or JAX (n=3), or PBS-gavaged (n=8) mice. Data were normalized using Gapdh. Similar experiments were performed three times. (C) Representative western blots of lysates from the EDTA-epithelial-rich fraction of different regions of the GI tract in animals with or without SFB colonization. (D–F) Role of SAAs in intestinal LP Th17 cell differentiation following SFB colonization. (D,E) IL-17A reporter littermates in WT (black, n=5) or Saa1/2 double knock-out (DKO) (grey, n=5) backgrounds were colonized with SFB for 7 days. RORγt+ cells among CD4+ LP T cells and their IL-17A producing capacity were measured by FACS. MFI and cell numbers of CD4+RORγt+Foxp3GFP+ Th17 cells were quantified. (F) Naïve CD4+ T cells from SFB-specific TCR transgenic (7B8) mice were adoptively transferred into SFB-colonized WT (circles, n=4) or Saa1/2 DKO (squares, n=4) littermates (as indicated in diagram). Total number of donor cells recovered from ileum (far left), percentage of RORγt-expressing donor-derived cells (center left), percentage of IL-17A–expressing cells upon ex vivo re-stimulation by PMA/Ionomycin (center right), and their total number (far right) were monitored by FACS at 7 days post transfer. N.S, not significant (P>0.05); *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001. Data are represented as mean with SD. See also Figure S2.
Figure 3. SAA1 promotes IL-17A production in polarized Th17 cells in vitro
(A) Schematic representation of the experimental protocol. CFSE-labeled naïve CD4+ T cells from WT mice were activated under non-polarizing (Th0: Anti-CD3 and Anti-CD28) or Th17 polarization condition (Th17: Anti-CD3, Anti-CD28, IL-6 and TGF-β). At 24h, RORγt+ T cells were further cultured with recombinant mouse SAA1 (rmSAA1) under Th17 polarization conditions for 48h. (B) IL-17A and IL-17F production by in vitro-polarized mouse Th17 cells (CD4+ RORγt+) at 48h after rmSAA1 (10 µg/ml) addition. Cells were re-stimulated with PMA/Ionomycin for 5h, and cytokine production and CFSE dilution were monitored by FACS analysis. (C) Representative data from polarized cells (48h) (4 experiments total). The frequencies of IL-17A- and IL-17F–producing cells during each cell division are displayed using the gates represented in (B). Data are shown as mean with SD. (D) RNAseq studies from two biological replicates comparing in vitro Th17 cells cultured in 20 ng/ml IL-6 and 0.1 ng/ml TGF-β with or without 2 µg/ml rmSAA1 for 48h. Dots represent FPKMs of individual mRNA transcripts in the respective cell type (grey). 195 genes that were significantly different between vehicle and SAA treatment (p-value<0.05, fold change greater than 2) are highlighted in black. (E) Ingenuity Pathway Analysis for upstream regulators of the 195 SAA-affected genes in Th17 cells. (F) A ROR-responsive element (RORE) firefly luciferase reporter was used to monitor RORγt transcriptional activity in Th17 cells. The RORE-luciferase reporter construct was introduced by nucleofection into Th17 cells polarized for 48h and cultured with or without rmSAA1 (2 µg/ml), and luciferase activity was measured 24h later. Data were normalized by renilla reporter activity. Results from four technical repeats in each condition were averaged. Results are representative of three independent experiments. *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001. See also Figure S3.
Figure 4. Requirement for IL-22 activation of epithelial Stat3 in SFB-dependent induction of SAA1/2
(A) Immunohistochemistry analysis of phospho-Stat3 (green) in terminal ileum from SFB-colonized (SFBe) or SFB free (JAX) mice on day 1 or day 3 following gavage. EpCAM (red) and DAPI (blue) staining was used to identify the intestinal epithelial layer. The experiment was repeated 2 times with similar results. (B) qRT-PCR analysis of SAA1/2 expression in the epithelial fraction of terminal ileum from Villin-Cre mice sufficient or deficient for epithelial Stat3 at 4 days after SFB colonization. Data were normalized using Gapdh. (C) Kinetics of SFB colonization post oral gavage of SFBe or JAX fecal contents. SFB genomic 16S in terminal ileum was quantified by qPCR analysis. Copy number of the SFB genomic16S was normalized to host Il-23r genomic DNA (JAX; n=4. SFBe; n=7). (D,E) Kinetics of IL-22 (D) and SAA1/2 (E) mRNA expression post SFBe gavage. (D) Level of IL-22 mRNA from isolated ileal lamina propria cells was assessed (JAX; n=7. SFBe; n=8 in each time points). (E) Level of SAA1/2 mRNA from epithelial fraction in ileum (JAX; n=4. SFBe; n=7). Graphs (C–E) show accumulated data from two or three independent experiments as mean with SD. Day 0 samples were from animals not gavaged with additional fecal samples. (F) SAA1/2 mRNA expression, determined by qRT-PCR, in epithelial fraction of terminal ileum from IL-22 sufficient or deficient mice at 4 days after SFB gavage (JAX gavaged: n=10, SFBe gavaged IL-22+/+ and +/− : n=5, SFBe gavaged IL-22−/− : n=5). (G) SAA1/2 mRNA expression in small intestinal epithelial cell organoid cultures following rIL-22 treatment. All qRT-PCR experiments were repeated two times with similar results, and data were normalized to GAPDH mRNA. Each dot represents a single mouse, and the orange bar indicates average relative gene expression of each group. N.S, not significant (P>0.05); *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001. See also Figure S4.
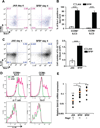
Figure 5. SFB colonization induces IL-22 production by ILC3s
(A) Cell surface expression of Sca-1 and CCR6 on ileal SILP ILC3s from mice at 4 days after oral-gavage with JAX or SFBe fecal contents. (B) Frequency of Sca-1-positive CCR6+ or CCR6− ILC3s in the terminal ileum from JAX- or SFBe-gavaged mice at 4 days after gavage. Data are represented as mean with SD (JAX gavaged: n=5, SFBe gavaged: n=6). (C) IL-22 production in ileal SILP ILC3. Ileal LP cells from JAX- or SFBe-gavaged mice were isolated at 4 days after gavage and cultured in vitro with GolgiStop (without PMA/Ionomycin re-stimulation) for 4h. IL-22-producing ILC3s were analyzed by FACS, and cell numbers were calculated (right panel). Bar graph shows accumulated data as mean with SD (JAX gavaged: n=5, SFBe gavaged: n=6). (D) IL-22 production in each indicated cell type isolated from the ileal LP of SFB-free (JAX; green line) and SFB-colonized (SFBe; magenta line) mice, analyzed at 4 days after gavage. (A–D) The experiment was repeated at least three times with similar results. (E) SAA1/2 expression, analyzed by qRT-PCR, in the epithelial fraction from terminal ileum of Rag2-sufficient or -deficient mice, at 3 days after SFB (SFBe) or control (JAX) gavage. Data are from two independent experiments. Each symbol represents a single mouse, and the orange bar indicates average of relative gene expression in each group. Experiments were repeated two times with similar results and data were normalized by GAPDH mRNA and combined. *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001. See also Figure S5.
Figure 6. IL-23R requirement for SFB-induced expression of IL-22 and SAA1/2 in ILC3 and epithelial cells, respectively
(A) IL-22 production in CCR6+ and CCR6− ileal LP ILC3s from _Il-23r_–sufficient or -deficient mice. (B) Cell number of IL-22-producing CCR6− (left) or CCR6+ (right) ileal LP ILC3s in JAX- or SFBe-gavaged mice, at 4 days after gavage. Bar graphs show accumulated data from two independent experiments with a total of 10 SFB-free (JAX) and 5 SFB-colonized (SFBe) mice of each genotype, shown as mean with SD. (C) SAA1/2 expression, analyzed by qRT-PCR, in the epithelial fraction from terminal ileum of indicated mice (JAX gavaged; n= 9, SFBe gavaged _Il-23r_G/+; n=8, SFBe gavaged Il-23rG/G; n=14).This experiment was repeated twice with similar results. Each dot represents a single mouse, and the orange bar indicates average relative gene expression of each group. *, P<0.05; **, P<0.01; ***, p<0.001; and ****, P<0.0001.
Comment in
- "A Sledgehammer Breaks Glass but Forges Steel": Bacteria Adhesion Shapes Gut Immunity.
Pedicord VA, Mucida D. Pedicord VA, et al. Cell. 2015 Oct 8;163(2):273-4. doi: 10.1016/j.cell.2015.09.040. Cell. 2015. PMID: 26451476
Similar articles
- Serum Amyloid A Proteins Induce Pathogenic Th17 Cells and Promote Inflammatory Disease.
Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, Lin WY, Yeung ST, Silva HM, Li D, Hine A, Loke P, Hudesman D, Martin JC, Kenigsberg E, Merad M, Khanna KM, Littman DR. Lee JY, et al. Cell. 2020 Jan 9;180(1):79-91.e16. doi: 10.1016/j.cell.2019.11.026. Epub 2019 Dec 19. Cell. 2020. PMID: 31866067 Free PMC article. - IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria.
Flannigan KL, Ngo VL, Geem D, Harusato A, Hirota SA, Parkos CA, Lukacs NW, Nusrat A, Gaboriau-Routhiau V, Cerf-Bensussan N, Gewirtz AT, Denning TL. Flannigan KL, et al. Mucosal Immunol. 2017 May;10(3):673-684. doi: 10.1038/mi.2016.80. Epub 2016 Sep 14. Mucosal Immunol. 2017. PMID: 27624780 Free PMC article. - Redundant cytokine requirement for intestinal microbiota-induced Th17 cell differentiation in draining lymph nodes.
Sano T, Kageyama T, Fang V, Kedmi R, Martinez CS, Talbot J, Chen A, Cabrera I, Gorshko O, Kurakake R, Yang Y, Ng C, Schwab SR, Littman DR. Sano T, et al. Cell Rep. 2021 Aug 24;36(8):109608. doi: 10.1016/j.celrep.2021.109608. Cell Rep. 2021. PMID: 34433045 Free PMC article. - Protective role of Th17 cells in pulmonary infection.
Rathore JS, Wang Y. Rathore JS, et al. Vaccine. 2016 Mar 18;34(13):1504-1514. doi: 10.1016/j.vaccine.2016.02.021. Epub 2016 Feb 13. Vaccine. 2016. PMID: 26878294 Review. - STAT3 and its activators in intestinal defense and mucosal homeostasis.
Hruz P, Dann SM, Eckmann L. Hruz P, et al. Curr Opin Gastroenterol. 2010 Mar;26(2):109-15. doi: 10.1097/MOG.0b013e3283365279. Curr Opin Gastroenterol. 2010. PMID: 20040863 Review.
Cited by
- Microbiota-induced plastic T cells enhance immune control of antigen-sharing tumors.
Najar TA, Hao Y, Hao Y, Romero-Meza G, Dolynuk A, Littman DR. Najar TA, et al. bioRxiv [Preprint]. 2024 Aug 14:2024.08.12.607605. doi: 10.1101/2024.08.12.607605. bioRxiv. 2024. PMID: 39185202 Free PMC article. Preprint. - Commensal microbiome and gastrointestinal mucosal immunity: Harmony and conflict with our closest neighbor.
Tian K, Jing D, Lan J, Lv M, Wang T. Tian K, et al. Immun Inflamm Dis. 2024 Jul;12(7):e1316. doi: 10.1002/iid3.1316. Immun Inflamm Dis. 2024. PMID: 39023417 Free PMC article. Review. - Nurturing gut health: role of m6A RNA methylation in upholding the intestinal barrier.
Wang S, Yang Y, Jiang X, Zheng X, Wei Q, Dai W, Zhang X. Wang S, et al. Cell Death Discov. 2024 Jun 3;10(1):271. doi: 10.1038/s41420-024-02043-x. Cell Death Discov. 2024. PMID: 38830900 Free PMC article. Review. - Antigen-level resolution of commensal-specific B cell responses can be enabled by phage display screening coupled with B cell tetramers.
Verma S, Dufort MJ, Olsen TM, Kimmel S, Labuda JC, Scharffenberger S, McGuire AT, Harrison OJ. Verma S, et al. Immunity. 2024 Jun 11;57(6):1428-1441.e8. doi: 10.1016/j.immuni.2024.04.014. Epub 2024 May 8. Immunity. 2024. PMID: 38723638 - IL-22, a vital cytokine in autoimmune diseases.
Li J, Wu Z, Wu Y, Hu X, Yang J, Zhu D, Wu M, Li X, Bentum-Ennin L, Wanglai H. Li J, et al. Clin Exp Immunol. 2024 Nov 12;218(3):242-263. doi: 10.1093/cei/uxae035. Clin Exp Immunol. 2024. PMID: 38651179 Review.
References
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. - PubMed
- Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI_/Howard Hughes Medical Institute/United States
- R01 DK103358/DK/NIDDK NIH HHS/United States
- T32 CA009161/CA/NCI NIH HHS/United States
- R01DK103358/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous