Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins - PubMed (original) (raw)
Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins
Yuan Lin et al. Mol Cell. 2015.
Abstract
Eukaryotic cells possess numerous dynamic membrane-less organelles, RNP granules, enriched in RNA and RNA-binding proteins containing disordered regions. We demonstrate that the disordered regions of key RNP granule components and the full-length granule protein hnRNPA1 can phase separate in vitro, producing dynamic liquid droplets. Phase separation is promoted by low salt concentrations or RNA. Over time, the droplets mature to more stable states, as assessed by slowed fluorescence recovery after photobleaching and resistance to salt. Maturation often coincides with formation of fibrous structures. Different disordered domains can co-assemble into phase-separated droplets. These biophysical properties demonstrate a plausible mechanism by which interactions between disordered regions, coupled with RNA binding, could contribute to RNP granule assembly in vivo through promoting phase separation. Progression from dynamic liquids to stable fibers may be regulated to produce cellular structures with diverse physiochemical properties and functions. Misregulation could contribute to diseases involving aberrant RNA granules.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
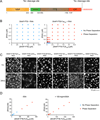
Figure 1. Particular IDRs Are Sufficient to Drive LLPS at Low Salt Concentration
(A) Schematic of SNAP-IDR proteins. MBP, maltose binding protein. SNAP, SNAP-tag used for fluorophore labeling. IDR, intrinsically disordered region. TEV protease removes MBP and His tags. (B) Fluorescence microscopy images of the macroscopic structures formed by SNAP-IDRs at 37.5 and 150 mM NaCl. SNAP-hnRNPA1IDR, 6.925 µM; SNAP-FusIDR, 10.75 µM; all the other proteins, 32.75 µM. Proteins were labeled with SNAP-Surface 649.
Figure 2. RNA Can Promote LLPS of IDR Proteins
(A) Schematic of SNAP-PTB-IDR proteins. PTB, polypyrimidine tract-binding protein, containing four RRM RNA binding domains. TEV protease removes MBP and His tags. (B) Phase diagram of SNAP-PTB and SNAP-PTB-FusIDR plus RNA. Red dots indicate phase separation; blue dots indicate no phase separation. (C) Fluorescence microscopy images of the macroscopic structures formed at 100mM NaCl by SNAP-PTB-4GIIIDR 1.25 µM and RNA 0.4 µM; SNAP-PTB-FusIDR 1.25 µM + RNA 0.4 µM; for the rest of droplets, SNAP-PTB-IDR 2.5 µM, RNA 0.8 µM. Proteins were labeled with SNAP-Surface 649. Images were taken at 1 hour and 24 hours after the initiation of phase separation by RNA addition. (D) Phase diagram of SNAP-PTB-FusIDR plus RNA in the absence and presence of 100 mg/ml bovine serum albumin (BSA).
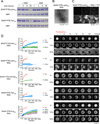
Figure 3. Phase Separated Droplets of SNAP-PTB-IDRs Plus RNA Mature Over Time
(A) SDS-PAGE of the high-salt soluble species present at different time points after the initiation of phase separation by RNA addition. MBP-SNAP-PTB-Lsm4IDR (5µM) or MBP-SNAP-PTB-TIA1IDR (5µM) were mixed with RNA (1.6µM) (phase separation) or buffer (no phase separation). At the indicated times, NaCl was raised to 500mM total concentration followed by 5 minutes of centrifugation, and the supernatant analyzed by SDS PAGE; gels were stained with coomassie blue. TEV protease was also present to remove MBP, which serves as an internal loading control. (B) Transmission electron micrographs of the high-salt insoluble species in a solution of SNAP-PTB-Lsm4IDR plus RNA after 24 hours incubation. (C) Representative images of increase over time in Thioflavin T (ThT) fluorescence for droplets of SNAP-PTB-Lsm4IDR(5 µM) plus RNA (1.6 µM) shown in the same intensity scale. (D) The liquid droplets of SNAP-PTB-IDR proteins plus RNA become less dynamic over time. Left panels show FRAP recovery curves. Data are reported as mean ± SD. Right panels show a representative droplet for each protein at different time points. See also Table S1.
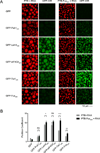
Figure 4. IDR Dependent Phase Separated Droplets Recruit Heterotypic IDRs
(A) Representative images showing the partitioning of GFP-IDR probes (100nM, green) into liquid droplets (red) of PTB or PTB-FusIDR plus Cy3-labeled RNA. (B) Quantification of the GFP-IDR partition coefficients in experiments from panel A. The partition coefficients are plotted as mean ± SD, from three independent measurements each of which averaged all the droplets across four random slide regions. ns. not significant, p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001. P values were determined by unpaired t test.
Figure 5. Full-length hnRNPA1 Undergoes IDR Dependent Phase Separation
(A) Schematic of domain architecture of SNAP-hnRNPA1 proteins. HRVC3 removes His and MBP tags. (B) Fluorescence microscopy images of the macroscopic structures formed by SNAP-hnRNPA1WT or SNAP-hnRNPA1ΔIDR at 37.5 mM NaCl, Images are shown in different intensity scale to highlight morphological changes. (C) Fluorescence microscopy images of structures formed by hnRNPA1WT, hnRNPA1ΔHexa, and hnRNPA1D262V (all 25 µM) at 37.5 mM NaCl. At indicated time points, NaCl was raised to 150 mM total concentration and structures that remained were imaged. Images are shown in different intensity scale to highlight morphological changes. (D) SDS-PAGE assays of the amount of high-salt soluble species present at different time points after the initiation of phase separation at 37.5 mM NaCl. hnRNPA1WT, hnRNPA1ΔHexa, and hnRNPA1D262V (all 25 µM) were incubated at 37.5 mM NaCl for the indicated time period before raising total NaCl concentration to 150 mM followed by centrifugation. The supernatant was then analyzed by SDS-PAGE with Coomassie staining. Quantification of the relative intensities of the bands is shown in the lower panel.
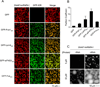
Figure 6. Phase Separated Droplets of Full-length hnRNP Al Recruit GFP-IDRs and Are Promoted by RNA
(A) Images showing the partitioning of GFP-IDR probes (100 nM, green) into the liquid droplets (red) of SNAP-hnRNP A1 (30 µM) at 37.5 mM NaCl. SNAP-hnRNPA1 was labeled with SNAP-Surface 649. (B) Quantification of the GFP-IDR partition coefficients in experiments from panel A. The partition coefficients are plotted as mean ± SD, from three measurements each of which averaged all the droplets across three slide regions. (C) Fluorescence microscopy images of SNAP-hnRNP A1 (2 or 20 µM) with or without RNA(2: 1 molar ratio of 5XA1 RNA : hnRNPA1) at 175 mM NaCl, 100 mg/ml PEG 3350.
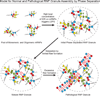
Figure 7. Possible Model for How Phase Separation Contributes to RNP Granule Assembly
Similar articles
- Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II.
Burke KA, Janke AM, Rhine CL, Fawzi NL. Burke KA, et al. Mol Cell. 2015 Oct 15;60(2):231-41. doi: 10.1016/j.molcel.2015.09.006. Epub 2015 Oct 8. Mol Cell. 2015. PMID: 26455390 Free PMC article. - Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization.
Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Molliex A, et al. Cell. 2015 Sep 24;163(1):123-33. doi: 10.1016/j.cell.2015.09.015. Cell. 2015. PMID: 26406374 Free PMC article. - Electrostatic modulation of hnRNPA1 low-complexity domain liquid-liquid phase separation and aggregation.
Tsoi PS, Quan MD, Choi KJ, Dao KM, Ferreon JC, Ferreon ACM. Tsoi PS, et al. Protein Sci. 2021 Jul;30(7):1408-1417. doi: 10.1002/pro.4108. Epub 2021 May 22. Protein Sci. 2021. PMID: 33982369 Free PMC article. - Multiple Modes of Protein-Protein Interactions Promote RNP Granule Assembly.
Mittag T, Parker R. Mittag T, et al. J Mol Biol. 2018 Nov 2;430(23):4636-4649. doi: 10.1016/j.jmb.2018.08.005. Epub 2018 Aug 9. J Mol Biol. 2018. PMID: 30099026 Free PMC article. Review. - Membraneless organelles: P granules in Caenorhabditis elegans.
Marnik EA, Updike DL. Marnik EA, et al. Traffic. 2019 Jun;20(6):373-379. doi: 10.1111/tra.12644. Epub 2019 Apr 11. Traffic. 2019. PMID: 30924287 Free PMC article. Review.
Cited by
- In vivo reconstitution finds multivalent RNA-RNA interactions as drivers of mesh-like condensates.
Ma W, Zhen G, Xie W, Mayr C. Ma W, et al. Elife. 2021 Mar 2;10:e64252. doi: 10.7554/eLife.64252. Elife. 2021. PMID: 33650968 Free PMC article. - A quantitative inventory of yeast P body proteins reveals principles of composition and specificity.
Xing W, Muhlrad D, Parker R, Rosen MK. Xing W, et al. Elife. 2020 Jun 19;9:e56525. doi: 10.7554/eLife.56525. Elife. 2020. PMID: 32553117 Free PMC article. - ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain.
Conicella AE, Zerze GH, Mittal J, Fawzi NL. Conicella AE, et al. Structure. 2016 Sep 6;24(9):1537-49. doi: 10.1016/j.str.2016.07.007. Epub 2016 Aug 18. Structure. 2016. PMID: 27545621 Free PMC article. - TRIP13 regulates progression of gastric cancer through stabilising the expression of DDX21.
Zhang G, Yang R, Wang B, Yan Q, Zhao P, Zhang J, Su W, Yang L, Cui H. Zhang G, et al. Cell Death Dis. 2024 Aug 26;15(8):622. doi: 10.1038/s41419-024-07012-x. Cell Death Dis. 2024. PMID: 39187490 Free PMC article. - Isolating and Analyzing Protein Containing Granules from Cells.
Victor RA, Thompson VF, Schwartz JC. Victor RA, et al. Curr Protoc. 2021 Mar;1(3):e35. doi: 10.1002/cpz1.35. Curr Protoc. 2021. PMID: 33740275 Free PMC article.
References
- Asherie N, Pande J, Lomakin A, Ogun O, Hanson SR, Smith JB, Benedek GB. Oligomerization and phase separation in globular protein solutions. Biophysical Chemistry. 1998;75:213–227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM063235/GM/NIGMS NIH HHS/United States
- R01 GM045443/GM/NIGMS NIH HHS/United States
- GM045443/GM/NIGMS NIH HHS/United States
- T32GM063235/GM/NIGMS NIH HHS/United States
- R37 GM045443/GM/NIGMS NIH HHS/United States
- R01-GM56322/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T32 GM065103/GM/NIGMS NIH HHS/United States
- R01 GM056322/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources